A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Applying Live Cell Imaging and Cryo-Electron Tomography to Resolve Spatiotemporal Features of the Legionella pneumophila Dot/Icm Secretion System
In This Article
Summary
Imaging of bacterial cells is an emerging systems biology approach focused on defining static and dynamic processes that dictate the function of large macromolecular machines. Here, integration of quantitative live cell imaging and cryo-electron tomography is used to study Legionella pneumophila type IV secretion system architecture and functions.
Abstract
The Dot/Icm secretion system of Legionella pneumophila is a complex type IV secretion system (T4SS) nanomachine that localizes at the bacterial pole and mediates the delivery of protein and DNA substrates to target cells, a process generally requiring direct cell-to-cell contact. We have recently solved the structure of the Dot/Icm apparatus by cryo-electron tomography (cryo-ET) and showed that it forms a cell envelope-spanning channel that connects to a cytoplasmic complex. Applying two complementary approaches that preserve the native structure of the specimen, fluorescent microscopy in living cells and cryo-ET, allows in situ visualization of proteins and assimilation of the stoichiometry and timing of production of each machine component relative to other Dot/Icm subunits. To investigate the requirements for polar positioning and to characterize dynamic features associated with T4SS machine biogenesis, we have fused a gene encoding superfolder green fluorescent protein to Dot/Icm ATPase genes at their native positions on the chromosome. The following method integrates quantitative fluorescence microscopy of living cells and cryo-ET to quantify polar localization, dynamics, and structure of these proteins in intact bacterial cells. Applying these approaches for studying the Legionella pneumophila T4SS is useful for characterizing the function of the Dot/Icm system and can be adapted to study a wide variety of bacterial pathogens that utilize the T4SS or other types of bacterial secretion complexes.
Introduction
Legionella pneumophila (L. pneumophila), the etiological agent of Legionnaires' disease, inhabits freshwater reservoirs, where the bacteria propagate by infecting and replicating within aquatic free-swimming protozoa. L. pneumophila causes disease outbreaks in humans when inhalation of aerosolized bacteria from potable water sources occurs. In infected cells, subversion of host pathways allows L. pneumophila to delay endocytic maturation of the vacuole in which it resides and to promote biogenesis of a cellular compartment that supports bacterial replication. This process is driven by a specialized bacterial type IVB secretion system (T4BSS) known as Dot/Icm and its repertoire of over 300 "effector" proteins that are translocated into the host cytosol during infection to facilitate manipulation of cellular functions1,2,3,4,5. Mutants lacking a functional Dot/Icm apparatus fail to deliver effectors into the host cytosol, are defective for intracellular replication, and are avirulent in animal models of disease6,7.
Many bacterial species have developed extremely complex and dynamic multicomponent machines that are required for infection processes. Other T4BSS like the Dot/Icm system are also essential for intracellular replication of bacterial pathogens such as Coxiella burnetii and Rickettsiella grylli. Although T4BSS are evolutionarily related to prototypical type IVA systems, which mediate DNA transfer and can deliver a limited repertoire of effector proteins, the Dot/Icm system has nearly twice as many machine components and delivers a wide variety of effectors. Presumably, this expansion in the number of components has enabled the Dot/Icm apparatus to accommodate and integrate new effectors easily8,9.
We recently used cryo-electron tomography (cryo-ET) to solve the structure of the Dot/Icm apparatus in situ and showed that it forms a cell envelope-spanning channel that connects to a cytoplasmic complex. Further analysis revealed that the cytosolic ATPase DotB associates with the Dot/Icm system at the L. pneumophila cell pole through interactions with the cytosolic ATPase DotO. We have discovered that DotB displays a cytosolic movement in most bacterial cells, indicating that this ATPase is present in a dynamic cytosolic population but also associates with the polar Dot/Icm complexes. In addition, DotO forms a hexameric assembly of DotO dimers associated with the inner membrane complex, and a DotB hexamer joins to the base of this cytoplasmic complex. The assembly of the DotB-DotO energy complex creates a cytoplasmic channel that directs the translocation of substrates through the T4SS (Figure 1)10.
Despite these recent advances, little is known about how the Dot/Icm system functions and how each protein assembles to form an active apparatus8. Uncovering the regulatory circuitry of the Dot/Icm T4SS is fundamental to understanding the molecular mechanisms of host-pathogen interactions. Therefore, we discuss how to use live cell microscopy and cryo-ET to detect and characterize essential L. pneumophila Dot/Icm system components that are tagged with super-folder GFP (sfGFP). Using quantitative fluorescence microscopy, the polar localization of DotB will be defined in a wild type background or when the type IV system is deleted. Time-lapse microscopy will be used to quantify differences in localization and dynamics between the Dot/Icm cytosolic ATPases.
The combined application of two complementary approaches such as live imaging and cryo-ET provides an advantage compared to other in vitro systems. Both methods are performed in intact cells and preserve the natural environment of the T4BSS, thus minimizing disruption of the native structure during sample preparation. Because overexpression of proteins may impair the stoichiometry of the secretion apparatus, sfGFP fusions are returned via allelic exchange to the Legionella chromosome so that each fusion is encoded in single copy and the expression is driven by the endogenous promoter. Visualization of chromosomally-encoded fusions enables quantification of the exact level of protein being expressed at a defined time point. Cryo-ET also has many advantages for determining the structure of secretion systems. The most notable advantage is that cryo-ET samples are comprised of frozen intact cells that preserve native complexes in the context of bacterial cell architecture. Consequently, cryo-ET may be preferable to biochemical purification approaches, which extract membrane complexes and may strip peripheral proteins from the core apparatus or modify the overall structure. In addition, tagging a protein of interest with a bulky protein such as sfGFP adds a mass that is detectable by cryo-ET and can assist with mapping the different subcomplexes of the Dot/Icm apparatus onto the structure obtained by cryo-ET.
This approach is a powerful tool for uncovering structural information about multimolecular complexes that assemble in the bacterial cell membrane. The interpretation of structures elucidated using these techniques will help the field understand how T4BSS components function, why so many components are required for function, how the components interact within the greater complex, and what functions these subassemblies perform.
Protocol
NOTE: All procedures involving the growth, manipulation, and imaging of L. pneumophila should be performed in a biological safety level 2 laboratory in compliance with local guidelines.
1. Insertion of sfGFP into L. pneumophila Chromosome Using Allelic Exchange and Double Selection Strategy (Figure 2, Figure 3)
- Clone into the gene replacement vector pSR47S11 the following sequence: the 1,000 bp upstream of the site of interest, then the sfGFP sequence, then the 1,000 bp downstream of the site of interest (Figure 2). The sfGFP sequence should be placed in frame to the N-terminus or C-terminus ends with a linker that contains four to eight amino acids. Transform the resulting vector into E. coli DH5αλpir. Later, streak L. pneumophila (the recipient) for single colonies on charcoal-yeast extract (CYE) agar12 containing 100 µg/mL streptomycin and grow for 5 days at 37 °C (Figure 3).
- Streak L. pneumophila on CYE-agar-streptomycin and grow for 2 days at 37 °C (heavy patch)10. Streak E. coli DH5α transformed with pRK600 helper plasmid (helper)13 on LB agar containing 25 µg/mL chloramphenicol. Streak the E. coli DH5αλpir (donor) on LB agar containing 50 µg/mL kanamycin.
- Perform triparental mating: incubate a colony of the helper, a colony of the donor, and the recipient by overlaying patches of the three strains on a CYE agar plate without selection and incubating for 4–8 h at 37 °C. As negative controls incubate a helper+recipient strain mix and a donor+recipient strain mix for the same periods of time.
- Resuspend the mating reactions in 500 µL of ddH2O. Plate 20 µL and 50 µL of the reactions on CYE agar containing 100 µg/mL streptomycin and 10 µg/mL kanamycin and grow for 5 days at 37 °C. Streak four of the resulting clones on CYE agar containing 100 µg/mL streptomycin and grow for 5 days at 37 °C.
- Streak 16 clones on CYE agar containing 5% sucrose and 100 µg/mL streptomycin and grow for 5 days at 37 °C. Then, streak 32 of these clones on CYE agar containing 100 µg/mL streptomycin and on CYE agar containing 100 µg/mL streptomycin and 10 µg/mL kanamycin and grow for 5 days at 37 °C.
2. Isolation of Clones that Integrated sfGFP into the L. pneumophila Chromosome
- Streak clones that were sensitive to kanamycin on CYE-agar-streptomycin plates and confirm the insertion of sfGFP into the chromosome with colony PCR. Use primers that are complementary to the sfGFP gene and to the chromosomal region of interest to amplify the insertion junction.
- Mix 0.5 µL of each of the 10 µM primers solutions and one colony to a final volume of 12.5 µL and denature for 10 min at 95 °C. Cool on ice for 10 min, add 12.5 µL of 2x PCR master mix solution, and perform a PCR analysis.
- Grow heavy patches of the isolated colonies on CYE-agar-streptomycin plates for 2 days at 37 °C. Examine the expression levels and stability of the sfGFP fusions by immunoblotting with an anti-GFP antibody.
3. Live Cell Imaging of L. pneumophila with Fluorescently Tagged Dot/Icm Components
- Preparation of agarose pads
- Make about 30 mL of a 1% low-melt agarose solution in water. Microwave in a glass flask for about 90 s, swirling occasionally, until the agarose is completely dissolved.
- Place two 22 x 22 x 0.15 mm3 glass slides on the edge of a 25 x 75 x 1.1 mm3 glass slide, one on top of the other. Stack two more 22 x 22 x 0.15 mm3 glass slides on the other edge.
- Pipette about 1 mL of the molten agarose into the center slide between the two upper glass slides, then place another 25 x 75 x 1.1 mm3 slide on top of the molten agarose. Try to avoid the formation of air bubbles. Cool the slides at 4 °C for 15 min.
- Using a scalpel or razor blade, gently cut the pad into small squares, ~5 x 5 mm2. Fix a double-sided adhesive 17 x 28 x 0.25 mm3 frame on a 25 x 75 x 1.1 mm3 glass slide and place several pads on the slide.
- Image acquisition
NOTE: The following steps are described for a microscope that is under the control of SlideBook 6.0 and equipped with solid state illuminators, CCD monochrome camera, and a 100x objective lens (1.4 numerical aperture). If needed, use alternative microscopy devices with appropriate hardware and software configurations that can be customized according to the protocol settings.- Dissolve a heavy patch of L. pneumophila in 1 mL of ddH2O, vortex and pipet 2–3 µL of the dilution onto the pads. Place a 50 x 24 x 0.15 mm3 coverslip gently over the adhesive frame.
- In the capture window adjust the ND to 180. Adjust the binning to 2×2 and use the 488 nm channel to expose the sample between 500–1,000 ms. Validate the specificity of the fluorescence signal by imaging untagged L. pneumophila with the same parameters (Figure 4).
4. Quantification of Polar Localization and Dynamics of Dot/Icm Components
NOTE: The following steps are designed for images with 0.129 µm per pixel that were acquired with 2 x 2 binning.
- Quantification of polarity for sfGFP fusion proteins (Figure 5)
- Adjust the image contrast so the bacteria are clearly visible. Use the region tool to place a 0.25 x 1.3 µm2 rectangle starting at the pole and extending into the cytoplasm. The rectangle must remain precisely within the bacterial borders.
- Mark at least 200 bacteria and use the Region to Mask button to create masks to the regions of interest. Under Mask Statistics and Mask Scope, choose Object. Then, under Features and Intensity, mark Mean Intensity and Variance.
- Export the data and calculate the polarity scores of each bacterium as the ratio between the variance to the mean intensity.
- For high-throughput applications use a phase objective and an appropriate condenser setup to acquire images with the phase and 488 nm channels. Make sure to choose fields of view where the bacteria are fully separated.
- Adjust the phase channel contrast of the image to a level where the bacteria are clearly visible. Open the dual channel image, launch the Create Segment Mask window and change the channel to phase.
- Adjust an appropriate threshold and remove small objects with the Define Objects button. Under Refine Mask choose Remove Edges Objects and later separate masks of bacteria that are adjacent to each other.
- Calculate the polarity scores of the signal in the 488 channel for each cell as was previously described in steps 4.1.2–4.1.3.
- Quantification of dynamics for sfGFP fusion proteins (Figure 6)
NOTE: Follow the instructions in section 3 to prepare a sample for image acquisition. Dynamics are defined as changes in intensity over time and the following steps are designed for short imaging periods (i.e., several minutes). Add to the pad the appropriate supplements if longer imaging periods are desired.- In the Image Capture window, mark Timelapse, enter the time of intervals in the interval box, and enter 2 in the # of Time Points box. Acquire two successive images of L. pneumophila expressing the fluorescent protein of interest.
- Adjust the image contrast until the cells are clearly visible. Follow Figure 6A and the descriptions below to place three different masks. Use the region tool to place a 0.25 x 0.25 µm square in the middle of at least 400 cells.
- Use the Region to Mask button to create a mask (mask 1) of the squares of interest. Create a new empty mask (mask 2) and use the pixel tool or the polygon tool to mark the entire cell area of at least 25 random cells, which will be used to calculate fluorescence bleaching. Create a new empty mask (mask 3) and use the large brush tool to mark areas between the cells, which will be used for background subtraction.
- Under Mask Statistics and Mask Scope, choose Object for mask 1 and mask 2. Then, under Features and Intensity, choose Mean Intensity and export the data of the two masks. For mask 3, export the mean intensity of the entire mask.
- Calculate the change in fluorescence intensity for each object in mask 1 using the following formulas:

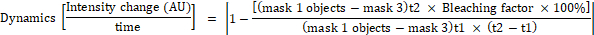
where mask 1 is a 0.25 µm × 0.25 µm square placed at the cell center, mask 2 covers the whole cell, mask 3 is the background between cells, t1 is the mean intensity in the first time point, and t2 is the mean intensity in the second time point.
5. Detection of sfGFP Mass Density with Cryo-ET
- Sample preparation, data collection, and reconstruction
- Grow a heavy patch of L. pneumophila expressing a fluorescently tagged Dot/Icm protein on CYE-agar-streptomycin plates for 48 hours at 37 °C. Resuspend the cells in ddH2O to OD600 ~0.7. Add 5 µL of colloidal gold particles (BSA Tracer, 10 nm) to 20 µL of the cell suspension.
- Pipette 5 μL of the cell mixture onto freshly glow-discharged holey carbon grid (R 2/1 on Cu 200 mesh) and let stand for 1 min. Blot with filter paper and freeze in liquid ethane using a gravity-driven plunger apparatus as described previously14,15.
- Image the frozen-hydrated specimens with a 300 kV transmission electron microscope equipped with a field emission gun, an energy filter, Volta phase plate, and a direct detection device. Collect single-axis tilt series at 26,000x and 42,000x magnifications, which result in pixel sizes at the specimen level of 5.4 Å/pixel or 3.4 Å/pixel, respectively.
- Use the tomographic package SerialEM to collect image stacks at ~0 μm defocus, with a range of tilt angles between -60° and +60° with 3° step increment and accumulative dose of ~60 e-/Å2,16. Align dose-fractionated movie images in each stack using MotionCor217. Assemble drift-corrected stacks using TOMOAUTO14.
- Align drift-corrected stacks by the IMOD marker-dependent alignment18. Reconstruct tomograms with the SIRT method19 for segmentations and direct image analysis and the WBP method20.
- Subtomogram analysis of sfGFP fusions samples
- Use the tomographic package I3 (0.9.9.3) for subtomogram analysis14,21,22.
NOTE: The alignment proceeds iteratively, with each iteration consisting of three parts in which references and classification masks are generated, subtomograms are aligned and classified, and class averages are aligned to each other. - Use 4 x 4 x 4 binned subtomograms for an initial alignment. Merge particles that belong to class averages and show electron density corresponding to sfGFP. After sorting particles with sfGFP fusions, use 2 x 2 x 2 binned subtomograms for a focused alignment of a region of interest (like the Dot/Icm cytoplasmic ATPase complex) to obtain a high-resolution structure.
- Use the tomographic package I3 (0.9.9.3) for subtomogram analysis14,21,22.
Results
Homologous recombination with double selection in two steps was used to construct the defined insertion of sfGFP. In the first step, triparental mating was performed, where the pRK600 conjugative plasmid (an IncP plasmid) from the E. coli helper strain MT616 was mobilized to the donor E. coli strain with the suicide vector pSR47S containing the sfGFP gene flanked by the two homologous regions, the origin of transfer oriT and the Bacillus subtilis counterselection gene sacB. Next, the c...
Discussion
Elucidating the functions of bacterial secretion systems is key to a complete understanding of host-pathogen interactions. Secretion systems are complex machines that can inject effectors proteins into host cells, and in some cases promote establishment of a subcellular niche that supports bacterial replication. The above method provides important new tools for studying the Dot/Icm secretion system of the respiratory bacterial pathogen Legionella pneumophila, yielding clues to the mechanisms of effector transloc...
Disclosures
The authors have nothing to disclose.
Acknowledgements
D.C. and C.R.R. were supported by the NIH (R37AI041699 and R21AI130671). D.P., B.H., and J.L were supported by the National Institutes of Health (R01AI087946 and R01GM107629).
Materials
| Name | Company | Catalog Number | Comments |
| 10 nm colloidal gold particles | Aurion | 25486 | |
| 100x Plan Apo objective (1.4 NA) | Nikon | ||
| ACES | Sigma-Aldrich | A9758 | |
| Activated charcoal | Sigma-Aldrich | C5510 | |
| Agaroze GPG/LMP, low melt | American bioanalytical | AB00981 | |
| Bacto dehydrated agar | BD | 214010 | |
| CoolSNAP EZ 20 MHz digital monochrome camera | Photometrics | ||
| Gene Frame, 1.7x2.8 cm, 125 µL | Fisher Scientific | AB-0578 | |
| Holey Carbon grid R 2/1 Cu 200 mesh | Quantifoil | Q225-CR1 | |
| Iron(III) nitrate nonahydrate | Sigma-Aldrich | 216828 | |
| K2 Summit camera for cryo-EM | GATAN | ||
| L-Cysteine | Sigma-Aldrich | C7352 | |
| Microscope cover slides 22x22 mm | Fisher Scientific | 12-542B | |
| Microscope cover slides 24x50 mm | Fisher Scientific | 12-545K | |
| Microscope slides 25x75x1 mm | Globe Scientific | 1380 | |
| SlideBook 6.0 | Intelligent Imaging Innovations | ||
| Spectra X light engine | Lumencor | ||
| Taq 2X Master Mix | New England BioLabs | M0270 | |
| Titan Krios | Thermo Fisher Scientific | ||
| Yeast Extract | BD | 212750 |
References
- Franco, I. S., Shuman, H. A., Charpentier, X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cellular Microbiology. 11, 1435-1443 (2009).
- Burstein, D., et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLOS Pathogens. 5, 1000508 (2009).
- Ninio, S., Roy, C. R. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends in Microbiology. 15, 372-380 (2007).
- Vogel, J. P., Andrews, H. L., Wong, S. K., Isberg, R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 279, 873-876 (1998).
- Isberg, R. R., O'Connor, T. J., Heidtman, M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nature Reviews Microbiology. 7, 13-24 (2009).
- Roy, C. R., Berger, K. H., Isberg, R. R. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within min of bacterial uptake. Molecular Microbiology. 28, 663-674 (1998).
- Archer, K. A., Roy, C. R. MyD88-Dependent Responses Involving Toll-Like Receptor 2 Are Important for Protection and Clearance of Legionella pneumophila in a Mouse Model of Legionnaires' Disease. Infection and Immunity. 74, 3325-3333 (2006).
- Nagai, H., Kubori, T. Type IVB Secretion Systems of Legionella and Other Gram-Negative Bacteria. Frontiers in Microbiology. 2, 136 (2011).
- Kubori, T., Nagai, H. The Type IVB secretion system: an enigmatic chimera. Current Opinion in Microbiology. 29, 22-29 (2016).
- Chetrit, D., Hu, B., Christie, P. J., Roy, C. R., Liu, J. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nature Microbiology. 3, 678-686 (2018).
- Merriam, J. J., Mathur, R., Maxfield-Boumil, R., Isberg, R. R. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infection and Immunity. 65, 2497-2501 (1997).
- Feeley, J. C., et al. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. Journal of Clinical Microbiology. 10, 437-441 (1979).
- Andrews, H. L., Vogel, J. P., Isberg, R. R. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infection and Immunity. 66, 950-958 (1998).
- Morado, D. R., Hu, B., Liu, J. Using Tomoauto: A Protocol for High-throughput Automated Cryo-electron Tomography. Journal of Visualized Experiments. (107), e53608 (2016).
- Hu, B., Lara-Tejero, M., Kong, Q., Galan, J. E., Liu, J. In Situ Molecular Architecture of the Salmonella Type III Secretion Machine. Cell. 168, 1065-1074 (2017).
- Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 152, 36-51 (2005).
- Zheng, S. Q., et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods. 14, 331-332 (2017).
- Kremer, J. R., Mastronarde, D. N., McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. Journal of Structural Biology. 116, 71-76 (1996).
- Gilbert, P. Iterative methods for the three-dimensional reconstruction of an object from projections. Journal of Theoretical Biology. 36, 105-117 (1972).
- Radermacher, M. Weighted Back-projection Methods. Electron Tomography. , 245-273 (2007).
- Winkler, H., et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. Journal of Structural Biology. 165, 64-77 (2009).
- Winkler, H., Taylor, K. A. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 106, 240-254 (2006).
- Prevost, M. S., Waksman, G. X-ray crystal structures of the type IVb secretion system DotB ATPases. Protein Science. 27, 1464-1475 (2018).
- Miklos, G. L., Rubin, G. M. The role of the genome project in determining gene function: insights from model organisms. Cell. 86, 521-529 (1996).
- Reyrat, J. M., Pelicic, V., Gicquel, B., Rappuoli, R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infection and Immunity. 66, 4011-4017 (1998).
- Yu, J. Single-Molecule Studies in Live Cells. Annual Review of Physical Chemistry. 67, 565-585 (2016).
- Stewart, P. L. Cryo-electron microscopy and cryo-electron tomography of nanoparticles. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 9, (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved