A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Culture of Brain Capillary Pericytes for Cytosolic Calcium Measurements and Calcium Imaging Studies
In This Article
Summary
Brain capillary pericytes are essential players in the regulation of blood-brain barrier properties and blood flow. This protocol describes how brain capillary pericytes can be isolated, cultured, characterized with respect to cell type and applied for investigations of intracellular calcium signaling with fluorescent probes.
Abstract
Pericytes are associated with endothelial cells and astrocytic endfeet in a structure known as the neurovascular unit (NVU). Brain capillary pericyte function is not fully known. Pericytes have been suggested to be involved in capillary development, regulation of endothelial barrier tightness and trancytosis activity, regulation of capillary tone and to play crucial roles in certain brain pathologies.
Pericytes are challenging to investigate in the intact brain due to the difficulties in visualizing processes in the brain parenchyma, as well as the close proximity to the other cells of the NVU. The present protocol describes a method for isolation and culture of primary bovine brain capillary pericytes and their following usage in calcium imaging studies, where effects of agonists involved in brain signaling and pathologies can be investigated. Cortical capillary fragments are allowed to attach to the bottom of culture flasks and, after 6 days, endothelial cells and pericytes have grown out from the capillary fragments. The endothelial cells are removed by gentle trypsinization and pericytes are cultured for 5 additional days before passaging.
Isolated pericytes are seeded in 96-well culture plates and loaded with the calcium indicator dye (Fura-2 acetoxymethyl (AM)) to allow for measurements of intracellular calcium levels in a plate reader setup. Alternatively, pericytes are seeded on coverslips and mounted in cell chambers. Following loading with the calcium indicator (Cal-520 AM), calcium live-imaging can be performed using confocal microscopy at an excitation wavelength of 488 nm and emission wavelength of 510-520 nm.
The method described here has been used to obtain the first intracellular calcium measurements from primary brain capillary pericytes, demonstrating that pericytes are stimulated via ATP and are able to contract in vitro.
Introduction
Brain capillary pericytes, together with endothelial cells and astrocytes, constitute the NVU1,2,3. The endothelial cells, which form the structural basis of the capillaries, form long cylindrical tubes with a diameter of 5-8 µm. The endothelial cells are sporadically covered with pericytes and surrounded by protrusions from astrocytes; the astrocyte endfeet.
The blood-brain barrier (BBB), situated at the brain capillaries, is the main site for exchange of nutrients, gases and waste products between the brain and the blood. The BBB also protects the brain from endogenous and exogenous neurotoxins and serves as a barrier for the delivery of a large number of drug compounds. The barrier function is a focus area, as well as an obstacle, for drug companies developing central nervous system (CNS) medicines. This has spurred a large interest in investigating the cells of the NVU in culture4. Brain astrocytes and endothelial cells have been cultured and characterized in a number of studies, whereas the studies and protocols for pericyte culture are sparse.
Previously published protocols have described generation of brain capillary pericyte cultures to some degree, using a range of different approaches such as immunopanning5, high- and low-glucose media6, fluorescent-activated cell sorting7, density gradient centrifugation8, etc. Although these methods seem sufficient to obtain cultures of pericytes, some are time consuming, cost expensive and the pericytes obtained might not be ideal due to the number of culture passages that can de-differentiate the pericytes9. Furthermore, the potential of cultured pericytes in in vitro signaling studies has been fairly unexplored until now.
The present work focuses on the generation of pericyte cultures from isolated bovine brain capillaries and the subsequent setup for measurements and imaging studies of changes in intracellular calcium, an important intracellular second messenger. We briefly describe the isolation of capillaries from cortical gray matter (for details see Helms et al.10) and the isolation and culture of pericytes in pure monoculture without contamination with endothelial or glial cells. We then provide a protocol for seeding of pericytes in 96-well plates and loading protocols for the calcium probe Fura-2 AM. Finally, we show how pericytes can be used in real-time confocal imaging in microscope culture chambers and describe the protocols for this.
Protocol
1. Preparation of buffers and solutions for cell culturing
- Prepare collagen stock solution by dissolving 5 mg of collagen IV from human placenta in 50 mL of PBS overnight at 4 °C. Aliquot the stock solution into 5 mL portions and store at -20 °C.
- Prepare fibronectin stock solution by dissolving 5 mg of fibronectin in 5 mL of sterile water overnight. Store the fibronectin stocks in aliquots of 500 µL at -20 °C. When thawing, add PBS to a final volume of 50 mL to prepare the work solution and store it at 4 °C.
- Prepare Dulbecco's Modified Eagle Medium (DMEM) complete medium by adding 50 mL of fetal bovine serum (FBS), 5 mL of MEM nonessential amino acids and 5 mL of penicillin/streptomycin (0.1 g/L streptomycin sulfate and 100,000 U/L penicillin G sodium) to 500 mL of DMEM.
- Prepare 5 mg/mL heparin stock solution by dissolving heparin sodium salt in PBS and pass it through a 0.2 µm filter for sterilization. Store the stock solution at 4 °C.
- Prepare growth medium (GM) immediately before use; mix 10 mL of DMEM-comp and 250 µL of heparin stock solution per T75-flask.
2. Isolation of capillaries from fresh bovine brain
NOTE: Bovine brain capillaries are isolated and cultured as previously described (Helms et al.10).
- Collect brains from calves, no older than 12 months of age, from a slaughterhouse and bring directly to the lab on ice.
- Remove the meninges and collect all gray matter from the brain using a scalpel. Identify the meninges as the film covering the brain and the gray matter by its gray color.
- Use a 40 mL Dounce tissue grinder to homogenize the gray matter in Dulbecco's Modified Eagle Medium (DMEM). Fill the slim part of the tissue grinder 1/5 with gray matter suspension and add DMEM until the slim part is filled.
- Separate the capillaries from free cells and smaller tissue pieces by filtration of the homogenate through a 160 µm nylon net filter. Flush the filters with DMEM-comp. Retrieve the capillaries and pool the suspensions into 50 mL centrifugation tubes.
- Resuspend the capillaries in DMEM-comp and add an enzyme mix of DNase I (170 U/mL), collagenase type III (200 U/mL) and trypsin (90 U/mL). Leave the suspension for 1 h in a 37 °C water bath for digestion of the capillaries.
- Run the suspension through a 200 µm mesh filter and resuspend in FBS with 10% dimethyl sulfoxide (DMSO). Freeze the capillaries overnight at -80 °C and move them to liquid nitrogen the day after for long term storage.
NOTE: The protocol can be paused here.
3. Seeding and culturing of bovine capillaries
- Day 0: Mix 0.7 mL of collagen IV stock with 6.3 mL of PBS. Add the solution to a T75-flask and leave the flask for 2 h at room temperature (RT) or leave it overnight at 4 °C.
- Remove the collagen solution from the flask and wash three times with PBS.
- Add 7 mL of fibronectin work solution and leave the flask for 30 min at RT. Then, remove the fibronectin solution and seed the capillaries immediately after.
- During the 30 min waiting time, thaw one vial of capillaries in a 37 °C water bath.
- When the capillaries are thawed, transfer immediately to a centrifugation tube with 30 mL of DMEM-comp and centrifuge for 5 min at 500 x g and RT. Remove DMEM-comp from the tube and re-suspend the capillary-pellet in 10 mL of fresh DMEM-comp.
- Transfer the 10 mL suspension to the coated T75-flask and leave the capillaries to adhere to the bottom of the flask for 4-6 h in a 37 °C incubator at 10% CO2.
NOTE: The cell growth rate is higher at 10% CO2 rather than the conventional 5% CO2. - After 4-6 h of incubation inspect the flask under a light microscope. Fractions of capillaries should now be attached to the bottom of the flask (Figure 1, day 0).
- Prepare GM and aspirate the DMEN-comp medium very careful from the capillaries and replace it with 10 mL of freshly made GM.
- Day 2: Remove GM from the capillaries and replace with 10 mL of freshly made GM. Cellular outgrowth from the capillaries should be visible under a light microscope at this point (Figure 1, day 2-3).
4. Isolation of primary pericytes from bovine brain capillaries
- Day 4: Inspect the capillaries under a light microscope.
NOTE: The flask should now be approximately 60-70% confluent to provide an appropriate amount of pericytes (Figure 1, day 4). If this is not the case; replace the GM with 10 mL of fresh medium and leave the flask in the incubator for another day. - Aspirate the medium and wash the cells gently in PBS.
- Add 2 mL of thawed Trypsin-EDTA for endothelial cells and leave the flask in the incubator for 1-3 min. Take out the flask frequently and observe with the microscope during this time period.
NOTE: The endothelial cells should round up and detach from the flask; pericytes should be visible as cells with a "ghost"-morphology and still be attached to the surface of the flask. This is a tricky and important step. It is essential to remove most endothelial cells to avoid contamination of the pericyte monoculture, but prolonged trypsinization can also detach the pericytes. The trypsinization time can vary slightly from time to time, and it is therefore of utmost importance to observe the flask frequently with the microscope during the treatment. - Gently tap the flask, when the endothelial cells have started to round up, to detach the loosened endothelial cells.
- To stop the trypsinization, add 10 mL of DMEM-comp to the flask. Flush the flask carefully a few times with the medium to remove the endothelial cells. Aspirate the endothelial cell suspension from the flask. The endothelial cells can now be used for other purposes.
- Add 10 mL of DMEM-comp to the flask. Look under the light microscope to assure the pericytes are still present and attached to the bottom. Put the flask back into the incubator to allow the pericyte-enriched culture to grow.
NOTE: It is important to observe the culture during the following days. If there is still a fair amount of endothelial cells growing another trypsin-treatment can be performed. - Allow the pericyte monoculture to grow with change of DMEM-comp. medium every second day. Check the growth of the cells under the light microscope (Figure 1, day 5-8).
5. Generation and storage of a monoculture of primary bovine pericytes
- Day 8-9: Inspect the capillaries under a light microscope
NOTE: The pericytes should now have reached 70-80% confluency and grow in islands in the flask (Figure 1, day 9). If the confluency of the pericytes is less than 70%, allow the cells to grow for another day. The pericytes will not form a complete monolayer as the endothelial cells would. - Aspirate DMEM-comp and wash the pericytes with 7 mL of PBS.
- Add 2 mL of trypsin-EDTA to the flask and leave it in the incubator for 2-3 min. Place the flask frequently under the light microscope to observe when the pericytes round up and detach from the flask. When the pericytes have started to round up, the flask can be gently tapped to detach the cells.
- Gently tap the flask, when the pericytes have started to round up, to detach the cells.
- Add 10 mL of DMEM-comp to the flask to stop the trypsinization process. Flush the flask a few times with the medium to help detach the last pericytes.
- Transfer the 12 mL cell suspension to a 50 mL centrifugation tube and fill up to 30 mL with DMEM-comp.
- Centrifuge the cell suspension for 5 min at 500 x g and RT. Aspirate the DMEM-comp. carefully without touching the cell pellet. Resuspend the cell pellet in 3 mL of FBS with 10% DMSO.
- Transfer the cell suspension into cryovials; add 1 mL to each, so there will be a total of 3 vials per T75-flask of pericytes. Freeze the pericytes at -80 °C overnight and move them to liquid nitrogen the day after for long term storage.
NOTE: Cells may be counted before freezing for a later estimate of survival percentage. The protocol can be paused here.
6. Setting up a pericyte monoculture for experiments
- Coat a T75-flask with collagen IV and fibronectin using the same procedure as mentioned in section 3.1-3.4.
- While the flask is being coated with fibronectin, thaw one vial of pericytes in a 37 °C water bath.
- Transfer the now thawed pericytes from the cryovial to a centrifugation tube with 30 mL of DMEM-comp. Centrifuge the cell suspension for 5 min at 500 x g, RT.
- Carefully aspirate the medium, leaving the cell pellet at the bottom of the tube. Re-suspend the pellet in 10 mL DMEM-comp.
- Collect and transfer the cell suspension to the coated flask. Leave the flask with pericytes to grow in a 37 °C incubator at 10% CO2.
- Every second day, refresh the medium with 10 mL of fresh DMEM-comp.
NOTE: After 5 days of growth, the pericytes should have reached approximately 80% confluency. If the confluency is less, leave the cells to grow for another day or two. The cells should now be ready for seeding for further experiments.
7. Seeding of pericytes in a coated 96-well plate
- Dilute collagen IV as described in step 3.1. Add 100 µL to each well in a 96-well plate and incubate for 2 h at RT or overnight at 4 °C.
- Aspirate the solution and wash the wells three times with PBS.
- Add 100 µL of diluted fibronectin to each well and incubate at RT for 30 min. Remove the fibronectin solution and use the plate immediately.
NOTE: Depending on how well the pericyte batch is growing, there should be enough cells for seeding two plates. - Take out the pericytes from the incubator and aspirate the medium. Wash the cells with PBS.
- Add 2 mL of trypsin-EDTA to the pericytes and follow same procedure as in step 5.3-5.6.
- Aspirate the medium, without harming the cell pellet and re-suspend the pellet in 1 mL of fresh DMEM-comp.
- Take out 12 µL of cell suspension and add to a counting chamber. Under the light microscope, count at least 3 of 3x3 grids and use the average cell count per grid.
- Use the equation below to calculate the volume of cell suspension that should be added to each well to seed 10.000 cells per well, in the 96-well plate.
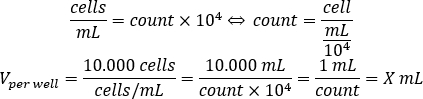
- Add DMEM-comp and the calculated volume of cell suspension in each well to reach a final volume of 200 µL.
- Place the 96-well plate in a 37 °C incubator at 10% CO2. Leave the cells to grow for 4 days with a change of medium after 2 days.
8. Preparation of buffers and solutions for Ca2+-imaging
- Autoclave coverslip cell chambers and coverslips.
- Assay buffer: Add 1.19 g of HEPES to 500 mL of HBSS buffer for a final concentration of 10 mM HEPES. Adjust the pH to 7.4.
- Prepare 20% (w/v) Pluronic F127 + 1% (v/v) polyethoxylated castor oil stock solution by dissolving 0.5 g of Pluronic F127 solution in 2.5 mL of anhydrous DMSO in a glass vial. Heat to 40 °C for approximately 30 min or until dissolved and vortex. Add 25 µL of polyethoxylated castor oil and store at RT. Do not freeze.
- Prepare 2 mM Fura-2 AM stock by dissolving 1 mg in 500 µL of anhydrous DMSO. Store in aliquots of 20 µL at -20 °C protected from light.
- Prepare 5 µM Fura-2 AM loading solution by mixing 20 µL of 20% Pluronic F-127 + 1% polyethoxylated castor oil stock solution with the 20 µL of 2 mM Fura-2 AM aliquot. Add 500 µL of assay buffer and vortex. Add assay buffer to a final volume of 8 mL. The solution should be prepared immediately before use and protected from light.
- Prepare 4 mM Cal-520 AM by dissolving 1 mg in 226.7 µL of anhydrous DMSO. Store in aliquots of 20 µL at -20 °C protected from light.
- Prepare 20 µM Cal-520 AM loading solution by mixing 20 µL of 20% Pluronic F-127 + 1% polyethoxylated castor oil stock solution with the 20 µL 4 mM Cal-520 aliquot. Add 500 µL of assay buffer and vortex. Add assay buffer to a final volume of 4 mL. The solution should be prepared immediately before use and protected from light.
9. Loading of pericytes with Fura-2 AM calcium indicator dye in a plate-reader setup
NOTE: All solutions should be at RT before the experiment starts.
- Take out the 96-well plate with cells from the incubator and aspirate the medium from the wells. Wash the cells twice with assay buffer.
- Add 100 µL of loading solution to each well and wrap the plate with tinfoil, to avoid photo bleaching. Incubate for 45 min with 30 rpm shaking at RT.
NOTE: Do not load Fura-2 AM at 37 °C, as it may load internal compartments. Remember to leave wells with cells in assay buffer instead of loading buffer; these are the "blanks" used for measuring background fluorescence. - Aspirate the loading buffer and wash the cells with assay buffer twice. Add 100 µL of fresh assay buffer and leave the cells to incubate for 30 min at RT; this allows for continuous cleavage of the AM-ester and thereby trapping Fura-2 AM inside the cells.
- Prior to the Ca2+-imaging, wash and replace the buffer with 100 µL of fresh assay buffer.
10. Well-plate fluorescence reading of pericytes in a plate-reader setup
- Set the temperature of the plate reader to 37 °C and transfer the 96-well plate with cells to the sample plate position. Place the reagent plate with agonist at the reagent plate position.
- Start by measuring loading of the cells to ensure equal loading of Fura-2 AM in all wells.
- Perform the measurements with excitation fluorescence wavelength at 340 nm/380 nm and the emission wavelength at 510 nm. Add 50 µL of agonist at speed 150 µL/s from the reagent plate to each well with cells in the sample plate position.
- Save the data and export as xlsx files for further analysis. Figure 2 shows the cytosolic Ca2+-response measured as the ratio between the two excitation wavelengths over time, where background fluorescence is subtracted.
NOTE: The plate-reader need to be a dual microplate reader with room for a "cell tray" and a "sample tray" and an integrated pipettor system.
11. Seeding of pericytes in a coated cell chamber for live imaging
NOTE: Coverslips may also be placed in the bottom of culture wells, coated and seeded with pericytes as described above, and then mounted in the chamber prior to experiments.
- Mount a coverslip into the cell chamber and make it tight to avoid leakiness.
- Dilute collagen IV as described in step 3.1. Add 500 µL to each cell chamber and incubate for 2 h at RT or overnight at 4 °C.
- Aspirate the collagen solution and wash the chambers three times with 500 µL of PBS.
- Add 500 µL of diluted fibronectin to each well and incubate at RT for 30 min. Remove the fibronectin solution and use the cell chamber straight afterwards.
- In the meantime, take out the flask with confluent pericytes and wash with 7 mL of PBS.
- Add 2 mL of trypsin-EDTA to the pericytes and follow same procedure as in step 5.3-5.6.
- Proceed by following the same steps as in step 8.6-8.7.
- Use the equation below to calculate the volume of cell suspension, which should be added to each chamber to seed 90.000 cells per chamber.
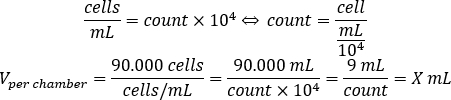
- Add DMEM-comp and the calculated volume of cell suspension in each chamber to reach a final volume of 500 µL.
- Place the cell chambers in the incubator at 37 °C, 10% CO2. Leave the cells to grow for 6 days (or until confluent).
NOTE: The pericytes grow slower on glass-slides compared to plastic; more days of growth are necessary.
12. Loading of pericytes with Cal-520 AM calcium indicator dye for live imaging
NOTE: All solutions should be at RT before the experiment starts.
- Prepare the 20 µM Cal-520 AM loading buffer: Mix 20 µL of 20% Pluronic F-127 + 1% polyethoxylated castor oil stock solution with the 20 µL 4 mM Cal-520 aliquot. Add 500 of µL assay buffer and vortex. Add assay buffer to a final volume of 4 mL. The solution should be prepared immediately before use and protected from light.
NOTE: Protect solutions containing Cal-520 AM from light exposure. - Take out the cell chambers from the incubator and aspirate the medium. Wash the cells twice with assay buffer.
- Add 500 µL of loading buffer to each chamber and incubate at RT for 45 min.
- Aspirate the loading buffer and wash the cells twice with assay buffer.
- Add 500 µL of fresh assay buffer to each chamber and incubate for 30 min at RT to allow cleavage of the AM-ester.
- Replace the buffer with 500 µL of fresh assay buffer before performing the live imaging at a confocal microscope.
13. Live imaging of intracellular Ca2+-levels
NOTE: A variety of microscope types can be used for the imaging. Upright or inverted conventional fluorescence microscopes, as well as upright or inverted confocal laser scanning microscopes with appropriate excitation source (488 nm) and emission filters (510-520 nm) can be used. Objectives should be suited for fluorescence and be of a high quality and with high numerical aperture (NA).
- Mount the cell chamber on the stage of the confocal microscope as gentle as possible, in order to avoid disturbance of the cells.
- Select an excitation wavelength of 488 nm, emission at 515 nm, sequential image acquisition with 5 second intervals, an XY image size of 512 x 512 pixels and measure for 2 min to measure baseline calcium signals.
- Add 3 µL of 100 mM ATP to the cell chamber with a pipette, and continue the sequential image acquisition. Perform the addition slowly and gently to not disturb the preparation and move the cells out of focus.
- Observe the degree of changes and increase the time interval over time as needed for approximately 18 min until no further morphological change is noted (Figure 3).
- Save time-lapse images and export them as TIFF and/or AVI files for further analysis.
NOTE: One vial of pericytes should give enough cells for seeding in 1-2 96-well plates and several coverslips, meaning you can prepare cells for both types of calcium-measurements.
Results
Bovine brain capillaries were isolated from fresh brain tissue and Figure 1 presents the capillary seeding and cellular outgrowth over days and subsequent purification of pericytes. The capillaries are fully attached to the flask at day 1 and on day 2 endothelial sprouting has become visible (Figure 1, day 2). After 4 days, the cellular outgrowth is highly distinctive (Figure 1, day 4a) and the ...
Discussion
In this study, we have presented a method to isolate primary pericytes from bovine brains. The described protocol allows culture of this otherwise rather inaccessible cell type. The subsequently obtained cell culture was a nearly homogenous population of pericytes, with little or no contamination with endothelial cells and glial cells based on cell morphology and protein expression12. Furthermore, we demonstrated a simple and straightforward method to load the pericytes with calcium dyes for ...
Disclosures
The authors declare no competing financial interests.
Acknowledgements
The authors wish to acknowledge funding from the Lundbeck Foundation Research initiative on Brain Barriers and Drug Delivery (RIBBDD) and Simon Hougners Family Foundation.
Materials
| Name | Company | Catalog Number | Comments |
| ATP | Tocris | 3245 | |
| Cal-520 AM | AAT Bioquest | 21130 | |
| Cell incubator | Thermo Fisher | ||
| Centrifuge | Thermo Fisher | Heraeus Multifuge 3SR+ | Standard large volume centrifuge for spinning down cells |
| Collagen IV | Sigma Aldrich | C5533 | |
| Confocal laser scanning microscope | Carl Zeiss | Zeiss LSM 510 | Inverted microscope |
| Counting chamber | FastRead | 102 | |
| Coverslip cell chamber | Airekacells | SC15022 | |
| Cremophor EL | Sigma Aldrich | C5135 | Formerly known as Kolliphor EL |
| DMSO | Sigma Aldrich | 471267 | |
| Dulbecco's Modified Eagles Medium | Sigma Aldrich | D0819 | |
| Fetal bovine serum (FBS) | PAA/GE Healthcare | A15-101 | |
| Fibronectin | Sigma Aldrich | F1141 | |
| Fura-2 AM | Thermo Fisher | F1201 | |
| Glass coverslips 22x22 mm | VWR International | 631-0123 | |
| HBSS | Gibco | 14065-049 | |
| Heparin | Sigma Aldrich | H3149 | |
| HEPES | AppliChem Panreac | A1069 | |
| Light microscope | Olympus | Olympus CK2 | Upright light microscope with phase contrast |
| MEM nonessential amino acids | Sigma Aldrich | M7145 | |
| Microplate Reader | BMG LabTech | NOVOstar | |
| PBS | Sigma Aldrich | D8537 | Phosphate-buffered saline |
| penicillin G sodium/streptomycin sulfate | Sigma Aldrich | P0781 | |
| Pluronic F127 | Sigma Aldrich | P2443 | |
| Trypsin-EDTA | Sigma Aldrich | T4299 | |
| T-75 flask | Sigma Aldrich | CLS3972 | |
| 96-well plate | Corning incorporated | 3603 |
References
- Armulik, A., Genove, G., Betsholtz, C. Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Developmental Cell. 21 (2), 193-215 (2011).
- Abbott, N. J., Ronnback, L., Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nature Reviews Neuroscience. 7 (1), 41-53 (2006).
- Mathiisen, T. M., Lehre, K. P., Danbolt, N. C., Ottersen, O. P. The Perivascular Astroglial Sheath Provides a Complete Covering of the Brain Microvessels: An Electron Microscopic 3D Reconstruction. Glia. 58 (9), 1094-1103 (2010).
- Helms, H. C., et al. In vitro models of the blood-brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. Journal of Cerebral Blood Flow and Metabolism. 36 (5), 862-890 (2016).
- Zhou, L., Sohet, F., Daneman, R. Purification of Pericytes from Rodent Optic Nerve by Immunopanning. Cold Spring Harbor Protocols. , 608-617 (2014).
- Liu, G. H., et al. Isolation, Purification, and Cultivation of Primary Retinal Microvascular Pericytes: A Novel Model Using Rats. Microcirculation. 21 (6), 478-489 (2014).
- Nayak, R. C., Herman, I. M. Bovine retinal microvascular pericytes: isolation, propagation, and identification. Methods In Molecular Medicine. 46, 247-263 (2001).
- Nees, S., et al. Isolation, bulk cultivation, and characterization of coronary microvascular pericytes: the second most frequent myocardial cell type in vitro. American Journal of Physiology-Heart and Circulatory Physiology. 302 (1), H69-H84 (2012).
- Armulik, A., Abramsson, A., Betsholtz, C. Endothelial/pericyte interactions. Circulation Research. 97 (6), 512-523 (2005).
- Helms, H. C., Brodin, B., Milner, R. Methods in Molecular Biology. Cerebral Angiogenesis: Methods and Protocols. 1135, 365-382 (2014).
- Abbracchio, M. P., Burnstock, G., Verkhratsky, A., Zimmermann, H. Purinergic signalling in the nervous system: an overview. Trends in Neurosciences. 32 (1), 19-29 (2009).
- Cai, C., et al. Stimulation-induced rises in cerebral blood flow and local capillary vasoconstriction depend on conducted vascular responses in brain capillaries. PNAS. , (2018).
- Tigges, U., Welser-Alves, J. V., Boroujerdi, A., Milner, R. A novel and simple method for culturing pericytes from mouse brain. Microvascular Research. 84 (1), 74-80 (2012).
- Maier, C. L., Shepherd, B. R., Yi, T., Pober, J. S. Explant Outgrowth, Propagation and Characterization of Human Pericytes. Microcirculation. 17 (5), 367-380 (2010).
- Orlidge, A., Damore, P. A. Cell specific effects of glycosaminoglycans on the attachment and proliferation of vascular wall components. Microvascular Research. 31 (1), 41-53 (1986).
- Rhinehart, K., Zhang, Z., Pallone, T. L. Ca2+ signaling and membrane potential in descending vasa recta pericytes and endothelia. American Journal of Physiology-Renal Physiology. 283, F852-F860 (2002).
- Bintig, W., et al. Purine receptors and Ca2+ signalling in the human blood-brain barrier endothelial cell line hCMEC/D3. Purinergic Signalling. 8 (1), 71-80 (2012).
- Neuhaus, A. A., Couch, Y., Sutherland, B. A., Buchan, A. M. Novel method to study pericyte contractility and responses to ischaemia in vitro using electrical impedance. Journal of Cerebral Blood Flow and Metabolism. 37 (6), 2013-2024 (2017).
- Kamouchi, M., et al. Calcium influx pathways in rat CNS pericytes. Molecular Brain Research. 126 (2), 114-120 (2004).
- Das, A., et al. ATP Causes Retinal Pericytes to Contract In vitro. Experimental Eye Research. 46, 349-362 (1988).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved