Method Article
Probing mRNA Kinetics in Space and Time in Escherichia coli using Two-Color Single-Molecule Fluorescence In Situ Hybridization
In This Article
Summary
This protocol describes an application of single-molecule fluorescence in situ hybridization (smFISH) to measure the in vivo kinetics of mRNA synthesis and degradation.
Abstract
Single-molecule fluorescence in situ hybridization (smFISH) allows for counting the absolute number of mRNAs in individual cells. Here, we describe an application of smFISH to measure the rates of transcription and mRNA degradation in Escherichia coli. As smFISH is based on fixed cells, we perform smFISH at multiple time points during a time-course experiment, i.e., when cells are undergoing synchronized changes upon induction or repression of gene expression. At each time point, sub-regions of an mRNA are spectrally distinguished to probe transcription elongation and premature termination. The outcome of this protocol also allows for analyzing intracellular localization of mRNAs and heterogeneity in mRNA copy numbers among cells. Using this protocol many samples (~50) can be processed within 8 h, like the amount of time needed for just a few samples. We discuss how to apply this protocol to study the transcription and degradation kinetics of different mRNAs in bacterial cells.
Introduction
The flow of genetic information from DNA to mRNA and protein is one of the most fundamental cellular processes, whose regulation is important for cellular fitness1. The number of mRNAs in a cell is determined by two dynamic processes, transcription, and mRNA degradation. However, how transcription and mRNA degradation are regulated in time and space of a single cell remains not completely understood, largely due to the shortage of experimental methods to quantitatively measure their kinetics in vivo.
Methods based on total mRNAs extracted from a population of cells, such as Northern blot, RT-PCR, RNA sequencing, and gene expression microarrays, can measure the relative difference in mRNA levels and have been widely used to analyze the rate of transcription elongation2,3,4,5 or the rate of mRNA degradation6,7. However, they do not provide the absolute number of mRNAs per cell, and hence, they are not suitable for probing the rate of transcription initiation8. Also, because mRNAs are extracted from a population of cells, the spatial distribution of mRNAs within a single cell and the variability of mRNA copy numbers among cells cannot be measured.
Next-generation RNA sequencing on individual cells (scRNAseq) can quantify the number of mRNAs per cell in a genomic scale9. However, it remains difficult to use this technique to measure transcription kinetics, due to challenges with sample preparation and high cost. In particular, the application of scRNAseq to bacteria has been technically difficult due to low mRNA abundance10,11.
Single-molecule fluorescence in situ hybridization (smFISH) is based on the hybridization of fluorescently-labeled single-stranded probes whose sequences are complementary to the target mRNA of interest12,13. The concept of sequence-specific hybridization is similar to that used in Northern blot or RT-PCR, but the hybridization is done in situ within fixed cells, to preserve the native localization of mRNAs. The signal of a single mRNA is amplified using many probes, ~20 nucleotides (nt) in length, hybridizing to different parts of an mRNA (Figure 1A)13. In this “tiling” probe approach, the number of probes needed to detect a single mRNA sets a lower limit on the length of mRNA that can be assayed. Alternatively, the mRNA of interest may be transcriptionally fused to a non-coding array of tandem Lac operator sequences, such that multiple copies of a fluorescently labeled lacO probe hybridize to a single mRNA (Figure 1B)14.
smFISH has been used to quantify the number of mRNAs per cell at steady state (i.e., when synthesis and decay are in balance) and to analyze the mean and variability of mRNAs among bacterial cells15,16,17. Recently, smFISH has been applied to quantify mRNA numbers at non-steady state, right after induction or repression of gene expression in E. coli18,19,20. The temporal changes in the absolute mRNA copy numbers were then used to calculate the rate of transcription initiation, elongation, and termination, as well as the rate of mRNA degradation. For this application, conventional smFISH procedures can be cumbersome because there are many samples, each representing one time point, that need to go through multiple buffer exchange steps (i.e., centrifugation and washing). Here, we describe an smFISH protocol, in which the sample handling steps are dramatically simplified by having cells adhered to the surface of a coverslip and by aspirating liquids with a vacuum filtration system14,19. Using the expression of lacZ in E. coli as an example, the full workflow (Figure 2) is demonstrated, including image analysis (Figure 3) yielding the in vivo kinetics of transcription (initiation, elongation, and termination) and mRNA degradation, cell-to-cell variability in mRNA expression, and mRNA localization. We anticipate that the protocol is widely applicable to probe in vivo kinetics and localization of other mRNAs in various bacteria species.
Protocol
1. Preparation of smFISH probes
NOTE: To label smFISH probes with a single fluorophore, follow a standard protocol for labeling nucleic acid oligonucleotides based on NHS ester chemistry21.
- Design smFISH probes. Decide whether to use “tiling” probes or “array” probes (Figure 1) for the gene of interest. See the Discussion section on how to make the decision.
- For “tiling” probes (Figure 1A), use an online probe designer tool (e.g., see Table of Materials).
- For “array” probes (Figure 1B), perform a BLAST sequence search to make sure that the probe sequence is not complementary to any other mRNA sequences.
- To study lacZ mRNA transcription and degradation kinetics, use two sets of 24 probes, each set covering the first and last 1 kb regions of lacZ (3,072 bp)19.
NOTE: These probe sets are, hereinafter, referred to as “5’ mRNA probe” and “3’ mRNA probe”, respectively. Sequences of these probes are listed in the Table of Materials.
- Order probe sequences as DNA oligonucleotides with a C6 amino linker at the 5’ end. Dissolve individual probes in water to 1 mM.
- Combine equimolar amounts of probes for “5’ mRNA probe” and “3’ mRNA probe” sets. For example, for the 5’ mRNA probe set for lacZ, combine 20 µL of each probe (total 24 kinds of probes in the set).
- Perform ethanol precipitation22 of the combined probes to remove any contaminations of primary and secondary amines (such as Tris, glycine, and ammonium salts) that can inhibit the conjugation reaction. In the end, dissolve the DNA pellet in 100 µL of water (yielding ~4.5 mM of DNA in a probe set).
NOTE: This step is recommended even if the probes underwent a standard desalt purification by the manufacturer. A standard filter-based purification may work in place of and in addition to the ethanol precipitation. - Choose two spectrally distinct fluorophores with a monofunctional NHS ester moiety, such that 5’ and 3’ mRNA probe sets can be labeled differentially. For example, prepare Cy5 NHS ester for 5’ mRNA probes and Cy3B NHS ester for 3’ mRNA probes. Dissolve each type of fluorophores in anhydrous DMSO to final 20 mg/mL (~25 mM).
- Prepare 0.1 M sodium bicarbonate (pH 8.5) right before each labeling reaction. Exposure to air for a long time will lower its pH and reduce the labeling efficiency.
- For the conjugation reaction, combine the following: 15 μL of the Cy5 fluorophore stock (from Step 1.5), 4 μL of 5’ mRNA probe set (from Step 1.4), 75 μL of sodium bicarbonate (from Step 1.6), and 7 μL of water. Wrap the tube with aluminum foil and shake at room temperature for 3-6 h.
NOTE: Longer incubation does not necessarily result in greater labeling efficiency. Also, the reaction can be scaled up or down if the concentrations of the components are maintained. - Repeat the above step for the 3’ mRNA probe set and the corresponding fluorophore (i.e., Cy3B NHS-ester).
- Perform ethanol precipitation22 to remove un-reacted dye molecules. Dissolve the pellet in ~50 μL of TE buffer (10mM Tris-HCl pH 8.0 with 1mM EDTA).
- Estimate the concentrations of DNA and fluorophore by using a UV-Vis spectrometer.
- Measure the absorbance at 260 nm and 559 nm (Cy3B) or 649 nm (Cy5). If the sample is too concentrated to yield an accurate measurement, dilute 1 µL of the sample to 10 µL.
- Convert the absorbance to the concentration:
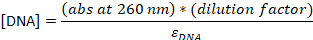
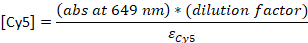
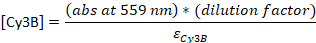
εDNA = 0.2 μM-1 (for 20-nt single-stranded DNA), εCy5 = 0.25 μM-1, and εCy3B = 0.13 μM-1
NOTE: [DNA] is the concentration of total probes within the solution. The concentration of individual probes is about 24x lower. The concentration of total probes will be used as “probe concentrations” from this point. If the ratio between [DNA] and [dye] is 1, the following HPLC step may be skipped23, and the sample should be diluted to final 4-5 μM in TE buffer.
- (Recommended) Purify the labeled probes from unlabeled probes and free dyes by using HPLC.
NOTE: Although this additional purification step will lead to the loss of sample, it is beneficial for the downstream applications. Removal of unlabeled DNA probes will increase the fluorescence signal from mRNA targets and removal of unreacted dyes will reduce background fluorescence.- Prepare HPLC with a standard analytical C18 column, 0.1 M triethylammonium acetate (TEAA) as buffer A, and acetonitrile as buffer B.
- Add 1 M TEAA to the sample (from Step 1.9) to make 0.1 M TEAA.
- Set the gradient program as follows: 0-5 min with 0% B, 5-35 min with a 0-30% linear gradient of B, 35-37 min with a 30-100% linear gradient of B, and 37-40 min with 0% B. Keep the flow rate at 0.1 mL/min and record chromatograms at 260 and 649 nm (for the Cy5-labeled samples) or at 260 and 559 nm (for the Cy3B-labeled samples).
- Collect the eluted sample when the absorbance increases in both DNA and fluorophore channels.
- Concentrate the eluted sample using a vacuum concentrator and re-suspend the pellet in 50-100 μL TE buffer.
- Check the concentration of DNA and fluorophore by using a UV-Vis spectrometer (see Step 1.10). Dilute, if necessary, to make final concentration around 4-5 µM. Store the probes at -20 °C
2. Preparation of solutions
- Prepare a large volume of DEPC-treated water and buffers (Table 1). These solutions can last over a year at room temperature.
- Prepare 4x fixing solution and wash solution (Table 1).
- Prepare the pre-hybridization solution and probe hybridization solution (Table 1). Prepare the probe hybridization solution during incubation in Step 5.1 or Step 6.1 and then keep the solution in a 37 °C countertop shaker for 20-40 min with a cover to minimize exposure to light.
NOTE: The concentrations of formamide, SSC, and probe were optimized for the lacZ probe sets to minimize background fluorescence while maximizing the real signal. See the Discussion section for details on how to modify these concentrations for different applications.
3. Preparation of coverslips and glass slides
- Clean coverslips and glass slides.
- Place individual coverslips and slides in a Coplin jar using forceps. Ensure that the coverslips and slides are separated and not touching each other.
- Fill the jar with 100% ethanol and close the lid. Place the jar in a water-bath ultrasonic cleaner and sonicate for 15-20 min.
NOTE: For the water-bath sonicator, it is recommended to turn off the heater function. - Pour out ethanol and wash with ultrapure water 3-4x. Use water flowing directly from the water purification machine.
- Pour out the water from the jar and fill it with 70% ethanol. Close the lid and perform sonication for 15-20 min and wash with ultrapure water.
- Fill the jar with ultrapure water and sonicate for 15-20 min.
NOTE: Coverslips and glass slides can be kept overnight in the Coplin jar filled with ultrapure water. - Take a slide or a coverslip out of the Coplin jar using clean forceps and blow-dry it using N2 gas. Repeat this for the remaining slides and coverslips.
- Place the dried slides in a clean storage box until use in Step 7.5. Place the dried coverslips in an empty 1,000-μL pipette tip box, which will serve as a “chamber” in the remaining procedure.
- Using a hydrophobic marker, draw circles on the coverslips following circular holes in the pipette tip box. These circles (~0.5 cm in diameter) will serve as “wells”. Wait at least 5-10 min for the marker to be completely dried.
NOTE: Always keep the lid of the tip box closed. - Apply a 20-μL drop of 0.1% poly-L-lysine to each well. Incubate for 10-50 min at room temperature.
NOTE: Adjust this volume according to the well size. Ensure that the solution completely covers the well area. For longer incubation, be careful to avoid evaporation. - After incubation aspirate poly-L-lysine without touching the surface as this will scrape the poly-L-lysine off. Then apply a drop (~20 μL) of DEPC water to the poly-L-lysine treated wells. Close the lid of the “chamber” to prevent evaporation until Step 5.1.
4. Time-course experiment and sample fixation
- Grow E. coli cells in ~20 mL liquid culture in a 250-mL flask. Keep the flask in a water bath shaker (30 °C) and continue shaking. Stop the shaker only when taking samples.
NOTE: Results presented in this paper are obtained from MG1655 cells grown in M9 minimal medium supplemented with 0.2% glycerol, 0.1% casamino acids, and 1 mg/L thiamine to an exponential growth phase (OD600~0.2). - Add 250 µL of the 4x fixing solution in an empty 1.5 mL tube. Repeat and prepare multiple tubes, as many as the time points to be taken. Label the tubes with time point numbers and keep them at room temperature.
- Take 750 µL of cell culture (OD600~0.2) before starting a time-course experiment. Add the culture to a tube marked for “time zero” (from Step 4.2). Invert the tube gently to mix cells with the fixing solution.
NOTE: Do not pipette up and down to mix, vortex, or “be rough” on the cells. This sample represents the repressed state and will be used as a control to calculate the fluorescence intensity of a single mRNA (see Step 9.4). - Add 0.02-1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) to the liquid culture to induce lacZ expression. Start a timer at this point (t = 0 min) and sample at a certain time interval (e.g., every 1 min) from then on. For sampling, repeat Step 4.3.
- Add 5 mM orthonitropheynl-β-D-fucopyranoside (ONPF) or 500 mM glucose24 at a certain time during the time-course experiment (e.g. at t = 1.5 min) to repress lacZ expression. After re-repression, continue to sample the cultures (Step 4.3) to track mRNA degradation.
NOTE: Repression can also be done with ~400 µg/mL rifampicin, a transcription initiation inhibitor25. - For fixation, incubate the tubes containing sampled cells at room temperature for 15 min, followed by incubation in ice for 30 min.
- To remove fixatives, centrifuge the tubes at 4,500 x g for 4 min at room temperature. Remove the supernatant with a pipette.
NOTE: Be sure to discard formaldehyde in a separate waste container following the safety protocol. - Add 1 mL DEPC-PBS and re-suspend the cells. Repeat centrifugation and re-suspension 2x more times.
NOTE: Fixed cells are fragile and need gentle treatment. Carefully re-suspend the pellet and avoid bubbles. - After the final wash step, re-suspend cells in ~30 µL DEPC-PBS.
5. Permeabilization of cell membranes
- Apply each time point sample to different wells on the coverslip (~30 μL per well). Wait for 10-30 min at room temperature for cells to adhere on the surface. Avoid merging of the liquid drops between wells.
- To rinse off unbound cells, aspirate the liquid and apply ~20 μL DEPC PBS to each well. Aspirate DEPC PBS within a few minutes.
- Permeabilize the cell membranes by applying 15 µL of 70% ethanol to each well for 4 min. Aspirate the ethanol after the 4 min, and make sure that the wells are completely dry.
NOTE: It is critical to limit the ethanol treatment for 4 min. Longer treatment will result in over-permeabilization. - Apply 30 µL of the wash solution to each well.
6. Probe hybridization
- Aspirate the wash solution from each well. Apply 30 µL of the pre-hybridization solution to each well. Incubate the chamber in the 37 °C oven for 30 min.
NOTE: Add ~50 mL of water to the bottom of the chamber to provide humidity. - Aspirate the pre-hybridization solution from each well. Apply ~30 µL of the probe hybridization solution to each well. Cover the chamber with aluminum foil and incubate in the 37 °C oven for 2 h.
NOTE: Make sure that the probe hybridization solution is in the 37 °C countertop shaker before this step. Avoid the merging of liquids between wells. Apply a smaller volume of the solution to each well, if needed.
7. Post-hybridization wash and preparation for imaging
- Using a multichannel pipette, apply ~30 µL of the wash solution to each well all at once. Aspirate and repeat 3-5x times of washing. Incubate the chamber in the 37 °C oven for 15-30 min.
- Repeat Step 7.1 two more times.
- Wash each well with DEPC-PBS 5x times. Follow the method used in Step 7.1 but skip the incubation process.
- Aspirate the liquid from the coverslip. Apply 4 µL of DEPC-PBS to each well.
- Using forceps, lift and flip the coverslip, and gently place it over a glass slide (from Step 3.2). Avoid bubbles.
- Seal the edges of the coverslip with silicone dental gum.
- Wait until the gum is solidified. One can pause here and store the slide overnight at 4 °C.
NOTE: Other smFISH protocols suggest adding oxygen scavenging reagents (e.g., glucose oxidase/catalase) or using a commercial anti-fade mounting medium14,26 to increase the photostability of the fluorophores.
8. Imaging
- To find an area of interest, use the live mode of phase contrast imaging. Change the field of view within a well by maneuvering the stage joystick. Choose an area where the cell density is optimal (i.e., there are many cells that are mostly separated). Adjust z-focus such that phase-contrast cell images are in focus.
- Take snapshots in the order of Cy5 (4-s exposure), Cy3 (2-s exposure), and phase contrast (0.2-s exposure).
NOTE: Cy3B dye molecules are imaged in the Cy3 channel, and the images are referred to as Cy3 images. - Repeat Steps 8.1-8.2 to acquire images of ~10 different areas within a well.
- Move the objective to another well and repeat Steps 8.1-8.3.
- Export images as TIFF files.
- (Optional) Image multi-color beads adsorbed on the coverslip surface in Cy5 and Cy3 channels to determine spatial shift between Cy5 and Cy3 channels for image registration purposes.
- Apply ~10 µL of multi-color fluorescent beads (0.2 µm diameter) on a clean coverslip surface and wait for 10-30 min. After washing with ~50 µL of PBS, apply ~5 µL of PBS and sandwich the coverslip with a glass slide. Seal and mount on the microscope.
- Image beads in both Cy5 and Cy3 channels.
9. Image analysis
NOTE: Matlab code used in this step is available in the following GitHub website: https://github.com/sjkimlab/Code_Publication/tree/master/JoVE_2020. The GitHub folder contains everything needed for the image analysis, including parameter values for cell segmentation and spot identification. The procedure in this step is further explained in the master script, called “FISHworkflow.m”.
- Open a cell segmentation tool, such as microbeTracker27 or Oufti28, and load phase contrast images. Choose “Independent frames” and press a button called “All frames” to begin the segmentation process, from which cells are identified and their contours are calculated (Figure 3B,C).
NOTE: Detailed protocols for using these software packages are available online (e.g., oufti.org). - Load Cy5 fluorescence images in the spotFinder function of microbeTracker or Oufti, and press the “Run” button to begin spot identification and quantification based on 2D Gaussian fitting (Figure 3B,C). Repeat this step for Cy3 fluorescence images to analyze spots in the Cy3 channel. This step produces a list of spots in each cell, including their intensities and coordinates.
- (Optional) Filter out dim spots (false positives) using a threshold, as explained in in the FISHworkflow.m file.
NOTE: Examine fluorescent spots in the negative control (e.g. MG1655 ΔlacZ) and determine the threshold to filter out false positives. - To obtain the spot intensity of a single mRNA, use a list of spot intensities measured at time zero (before adding IPTG), and fit the distribution of spot intensities with a Gaussian mixture model with two mixture components. Take the peak position of the first Gaussian population (black line in Figure 3D,E) as the spot intensity of a single mRNA. Perform this for Cy5 spots and Cy3 spots separately to obtain the spot intensity of a single 5’ and 3’ lacZ mRNA.
NOTE: Repeat this in every time-course experiment because the spot intensity of a single mRNA can vary slightly in different experiments. - Divide the fluorescence intensity of a spot with the intensity of a single mRNA (from Step 9.4) to obtain the number of mRNAs within a spot. Sum normalized spot intensities within a cell to calculate the total number of mRNA in a cell (Figure 3F). Perform these calculations for 5’ and 3’ mRNA separately.
- Calculate and plot the mean mRNA numbers per cell at each time point (e.g., Figure 4B), and analyze the in vivo kinetics of transcription and mRNA degradation from the temporal change in the mean mRNA levels (Figure 4B).
- To obtain the rate of transcription elongation, perform a least-squares fitting of a line to the initial rise in 5’ and 3’ mRNA signals and identify intercepts to the basal levels (Figure 4B). The difference between these intercepts indicates the average time for RNAPs to travel from the 5’ probe region to the 3’ probe region. Divide the distance between two probe sets (2 kb) with this time to obtain the average rate of transcription elongation.
- To obtain the rate of mRNA degradation, fit an exponential decay function, y = A·exp(-t/τ) to the final decay region of the 5’ and 3’ mRNA signals (e.g., Figure 4B). The fitting parameter, τ, is the average mRNA lifetime.
- (Optional) Analyze the cell-to-cell variation in gene expression (e.g., the cell-level response to the induction shown in Figure 4C), based on the distribution of mRNA numbers in each cell (calculated in Step 9.5).
- (Optional) Using information about spot location along the major and minor axes of a cell (obtained from Step 9.2), analyze the localization of mRNAs (Figure 4D,E).
- (Optional) Analyze co-localization of 5’ and 3’ mRNAs (Figure 5) by comparing localization of spots detected in the Cy5 and Cy3 channels.
- Load images of multi-color beads (Step 8.6) in the spotFinderF function in microbeTracker and obtain coordinates of bead centroids in Cy5 and Cy3 channels. Use the list of centroid coordinates to calculate the affine transformation matrix, which informs how Cy5 and Cy3 channels are shifted and rotated with respect to each other29.
- Apply the affine transformation matrix to Cy5 and Cy3 FISH images to convert Cy3 images in the Cy5 coordinate. Classify if a spot is co-localized with another spot in a different channel. For example, a spot in the Cy5 channel is considered to be co-localized with another spot in the Cy3 channel if the distance between their centroids is less than 150 nm (Figure 5).
- Analyze how many Cy5 spots are classified as “co-localized” with Cy3 spots at each time point. Also, analyze the intensity of the co-localized spots (Figure 5).
Results
Figure 3 shows representative images from this smFISH protocol. A full field of view (86.7 μm x 66.0 μm using our microscopy setup detailed in Table of Materials) shows ~500 E. coli cells dispersed throughout the field (Figure 3A). If the density of cells is much higher than what is shown in this image, automatic cell segmentation becomes difficult as segmentation algorithms do not reliably identify individual cells when cells touch each other. One needs to adjust the concentration of cells and incubation time used for surface adherence (Step 5.1) to achieve the optimal density of cells in the field of view.
The morphology of cells in the phase contrast images should remain comparable to that of live cells for segmentation purposes (Figure 3A-C). If cells are over-permeabilized, the cell morphology changes (like “ghosts”; Supplementary Figure 1). In that case, one may reduce the duration of 70% ethanol treatment in Step 5.3.
Before induction the average lacZ expression level was ~0.03 mRNAs per cell, consistent with previous reports15,30. Also, the distribution of lacZ mRNA spot intensities before induction did not fit well with a normal distribution or a Poisson distribution due to the presence of spots with high intensities (Figure 3D,E). This suggests that most of the spots detected under the repressed state represent a single lacZ mRNA, but a small population of spots contains more than one lacZ mRNA. To isolate the population with a single lacZ mRNA, we used a Gaussian mixture model with two mixture components (insets in Figure 3D,E). Then, the mean of the first Gaussian was taken as the mean intensity of a single mRNA spot (e.g., the peak of the black curve in Figure 3D) and used to convert the spot intensity to the number of mRNAs, for any spots detected in the time-course experiment. To calculate the total number of mRNAs within a cell, the normalized spot intensities were summed in each cell (Figure 3F)19.
When the expression level of lacZ mRNA is low, there are one or two diffraction-limited lacZ mRNA spots spatially separated within a cell. Hence, the images of these spots can be analyzed by 2D Gaussian fitting for their intensity and localization.
When the expression level is high, such that spots overlap with each other within a cell, 2D Gaussian fitting does not do reliable quantification. In that case, the mRNA level should be calculated by dividing the total, background-subtracted fluorescence signal within a cell with the mean intensity of a single mRNA19.
When the expression of lacZ is induced, the signal of 5’ lacZ mRNA increases first and that of 3’ lacZ mRNA increases later (Figure 4B). If the expression of lacZ is repressed, both 5’ and 3’ lacZ mRNA signals decrease with some delay in between (Figure 4B). To obtain the rate of transcription elongation, the rise of 5’ and 3’ signals are first fit with lines (Figure 4B), and the difference in x-intercepts are taken as the time for RNAPs to travel the distance between two probe regions (2,000 nt). The rate of transcription elongation can be measured from each time-course experiment and standard deviations can be calculated from experimental duplicates. The average rate of transcription elongation was 15-30 nt/s under our experimental conditions19.
Additionally, the rate of mRNA degradation (inverse of the mean mRNA lifetime) was obtained by fitting the decay region with an exponential function (Figure 4B). Our time-course data contains mRNA degradation during and after transcription31. We fit the time points after 3’ mRNA started to decay (t > 6 min) to probe the degradation of released mRNAs. We obtained ~90 s as an average lifetime of either 5’ or 3’ lacZ mRNA19.
The rate of transcription initiation can be calculated from the slope of 5’ signal increase after induction (Figure 4B, blue), or from the average mRNA number at steady state (which is the initiation rate divided by the degradation rate). Furthermore, the probability of premature transcription termination can be estimated, either by taking the ratio between the slope of 3’ signal increase vs that of 5’ signal increase32 or between the steady-state levels of 3’ and 5’ mRNA regions19.
Because smFISH is a single-cell technique, we can analyze cell-to-cell variability in transcription. For example, one can analyze the percentage of cells expressing lacZ mRNA after IPTG is added (Figure 4C). One can also address whether mRNA localization changes after induction. We observed that 5’ and 3’ lacZ mRNA spots move slightly outward, away from the center of the cell (Figure 4D,E), consistent with a previous report33.
Lastly, analysis of co-localization between 5’ and 3’ mRNA spots can be informative (Figure 5A). For example, in the repressed state (time zero), about 25% of 5’ mRNA spots are co-localized with a 3’ mRNA spot. At t = 1 min, as many gene loci have 5’ mRNA synthesis, but not yet 3’ mRNA synthesis, most of the 5’ mRNA spots are by themselves without 3’ mRNA signal (i.e., low probability of co-localization). However, when the 3’ mRNA appears (i.e., t = 2 min), the probability of co-localization increases (purple arrow in Figure 5A,B). This time point, when the co-localization becomes frequent, depends on the rate of transcription elongation. The 2-D density plot of 5’ and 3’ lacZ mRNA numbers within each co-localization spot detected at this time point can be used to infer the density of RNAPs on the lacZ gene (Figure 5C). As previously reported19, the 5’ mRNA numbers in this plot indicate that most of the lacZ loci have less than 10 RNAPs on the DNA when lacZ expression is induced by 1 mM IPTG. Additionally, the 3’ mRNA numbers in this plot is related to the clustering of RNAPs34. The fact that the number of 3’ mRNA is close to one means that roughly only one RNAP enters the 3’ probe region. This suggests that RNAPs on the lacZ gene are spatially separated, instead of forming a cluster (or “convoy”).
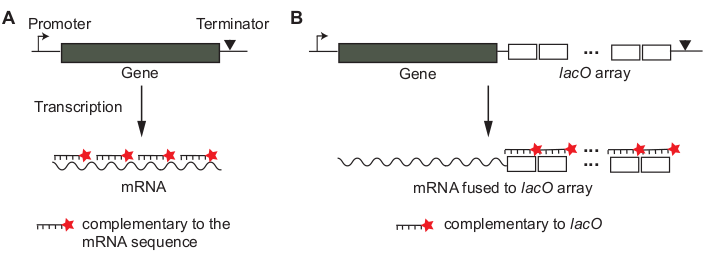
Figure 1: Design of smFISH probes for an mRNA of interest. (A) A tiling method. Sequences of short DNA oligonucleotides (~20 bp in length) are chosen so that they can cover the mRNA of interest. The oligonucleotide probes are labeled with a fluorescent dye molecule. (B) An array method. A non-coding array of tandem sequences (e.g., “lacO array”) is transcriptionally fused to the mRNA of interest. Fluorescently labeled probe complementary to the repeat unit (e.g., lacO probe of 17 bp in length) is used to amplify the signal of an mRNA. Please click here to view a larger version of this figure.
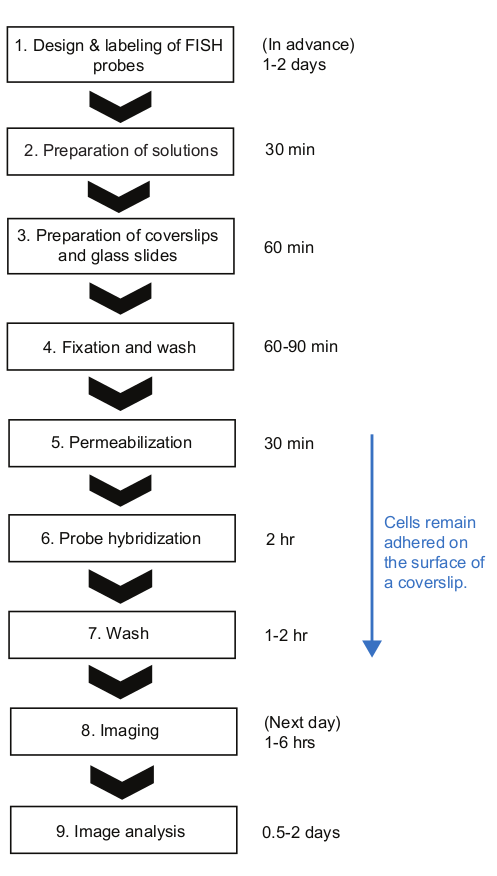
Figure 2: Schematic of smFISH experimental procedure and time duration of each step. Please click here to view a larger version of this figure.

Figure 3: smFISH image analysis. (A-C) smFISH microscopy image of 5’ lacZ mRNA (red) and 3’ lacZ mRNA (green) in wild-type E. coli (MG1655) grown in M9 minimal medium supplemented with 0.2% glycerol, 0.1% casamino acids, and 1 mg/L thiamine at 30 °C. (A) A representative image of a sample from t = 3 min after induction with 0.05 mM IPTG at t = 0 min and repression with 500 mM glucose at t = 1.5 min. Phase contrast and two fluorescence images of Cy5 (for 5’ lacZ mRNA, red) and Cy3 (for 3’ lacZ mRNA, green) were overlaid with pseudo-coloring. The image shows an entire field of 86.7 μm x 66.0 μm. Scale bar, 5 μm. (B) Zoom-in version of a small region (yellow box) in (A). Cell outlines are shown in white, and fluorescence spots identified from image analysis are shown with red dots. Scale bar, 1 μm. (C) Detection of cell outlines and fluorescent spots under a high expression condition (t = 4 min after induction with 1 mM IPTG). Scale bar, 1 μm. (D-E) Distributions of 5’ and 3’ mRNA spot intensities measured before adding IPTG (the repressed state). The histograms are shown with two Gaussian functions (black and grey) whose mean values are from the Gaussian mixture model. Inset shows quantile-quantile plot of random numbers generated from the Gaussian mixture models and experimentally measured mRNA spot intensities (n = 1040 for 5’ mRNA and 680 for 3’ mRNA). (F) Information obtained for an individual cell pointed in panel (B). For a given cell (i), spots were identified in Cy5 and Cy3 channels, and their intensity (I) and coordinate along the short and long axis of a cell (d, l) were quantified from 2D Gaussian fitting. After normalization spot intensities were summed to yield the total number of 5’ or 3’ mRNAs in this cell. Also, co-localization between spots from different channels can be analyzed as in the example shown in Figure 5. Please click here to view a larger version of this figure.

Figure 4: Analysis of in vivo kinetics of transcription and mRNA degradation. (A) Schematic and representative images of two-color smFISH experiments measuring changes in lacZ mRNA levels over time. Red and green dotted lines indicate Cy5 or Cy3B labeled oligonucleotide probes that hybridize to the 1-kb-long 5’ and 3’ mRNA regions of lacZ mRNA in E. coli, respectively. Also shown are overlays of two fluorescence images with a phase contrast image at indicated time points after induction with 0.2 mM IPTG at t = 0 min. Transcription was repressed with 500 mM glucose at t = 1.5 min. Scale bar, 1 μm. The figure has been modified from Kim et al19. (B) 5’ and 3’ lacZ mRNA numbers per cell over time, during the experiment described in the panel (A). Error bars are bootstrapped SEMs. At least 1,200 cells were analyzed per time point. The initial rise of the 5’ and 3’ mRNA signals was fit with a line (blue). The difference in x-intercepts was 1.93 min, yielding the average rate of transcription elongation of 17.3 nt/s. Final decay of the 5’ and 3’ mRNA signals was fit with an exponential decay function (grey). The fit parameters indicate that the average mRNA lifetime is 1.52 min for 5’ mRNA and 1.66 min for 3’ mRNA. (C) Percentage of cells with one or more lacZ mRNA spots during the experiment described in (A). Error bars are bootstrapped SEMs. (D) Localization of a spot along a cell’s short axis. One can quantify a spot’s proximity to the membrane by dividing the location along the short axis (d) with half width of the cell (w). (E) Change in the localization of 5’ and 3’ lacZ mRNA spots along cells’ short axis during the experiment described in (A). Please click here to view a larger version of this figure.
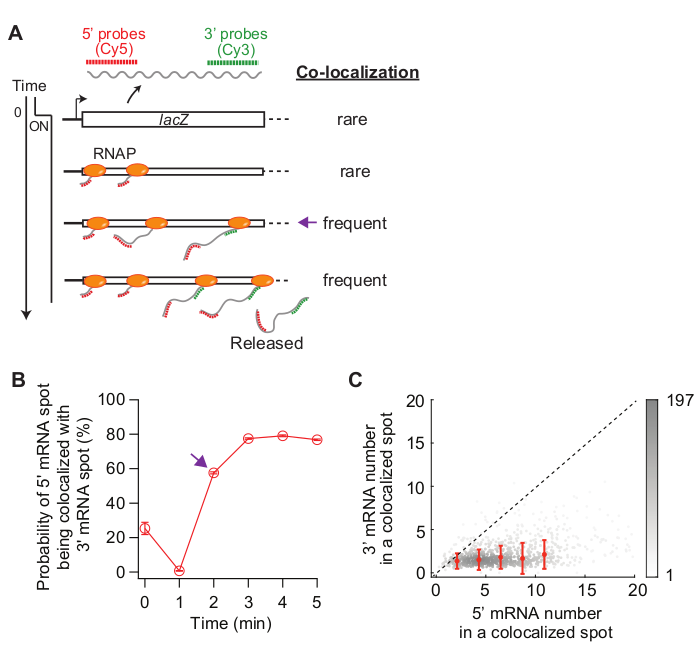
Figure 5: Analysis of co-localization of 5’ and 3’ mRNA spots. (A) Schematic showing the expected co-localization between 5’ and 3’ mRNA spots after induction. When 3’ mRNA is made, the probability of a 5’ mRNA spot being co-localized with a 3’ mRNA spot increases (purple arrow). (B) The probability of co-localization after induction with 1 mM IPTG. The purple arrow indicates the time point where the probability of co-localization first becomes frequent according to the schematic in panel (A). (C) The number of 5’ and 3’ lacZ mRNAs within a co-localization spot detected at t = 2 min after induction with 1 mM IPTG (total 841 spots). Gray dots represent individual co-localized spots, whereas red dots represent the average of binned data. Error bars are SEM. The shade of gray indicates the density of points in a given area of the graph. The dotted line indicates a slope of 1. Please click here to view a larger version of this figure.
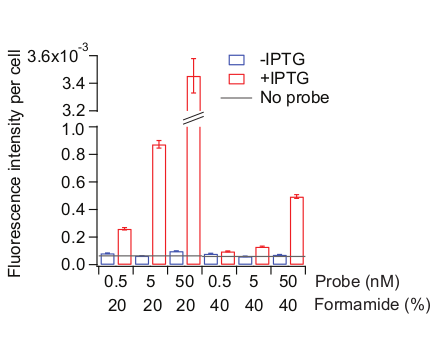
Figure 6: Optimization of the probe hybridization condition. Two kinds of samples were used: MG1655 cells grown as described in Figure 3 and remain uninduced (blue) or treated with 0.5 mM IPTG for 20 min (red). Probe hybridization solution was made with different concentrations of probes (total 72 Cy5-conjugated probes tiling the entire lacZ region) and of formamide. Formamide concentrations were also adjusted in the pre-hybridization solution and the wash solution, accordingly. “No probe” (grey line) indicates the fluorescence level of the IPTG-added cells treated with no probes during the hybridization step. Mean fluorescence intensity normalized by cell area (AU) was calculated from 300-800 cells. Error bars are bootstrapped SEMs. Please click here to view a larger version of this figure.
Supplementary Figure 1: Distorted cell morphologies due to over permeabilization. Overlay of phase contrast (gray scale), 5’ lacZ mRNA (Cy5, red), and 3’ lacZ mRNA (Cy3, green) images of MG1655 cells 5 min after the induction with 1 mM IPTG. (A) An example showing mixture of normal cells and overly permeabilized cells lacking normal morphology (indicated with pink arrows). (B) An example showing “ghosty” cells clumped together. Scale bar = 1 µm. Please click here to download this file.
| DEPC Water | ||||
| Add 0.1% DEPC to ultrapure water and incubate the bottle (covered) in the 37°C oven overnight and autoclave next day. | ||||
| DEPC PBS (10X) | ||||
| Mix the following: | ||||
| 80 g | NaCl (final 1.37 M) | |||
| 2 g | KCl (final 27 mM) | |||
| 14.2 g | Na2HPO4 (final 100 mM) | |||
| 2.7 g | KH2PO4 (final 20 mM) | |||
| Ultrapure water to 1L | ||||
| Filter (0.22 μm) into a glass bottle. | ||||
| Add 0.1% DEPC and follow the instruction for DEPC water. | ||||
| To make 1X solution, dilute 10 times with DEPC water. | ||||
| 1M DEPC sodium phosphate buffer, pH 7.4 | ||||
| Mix the following: | ||||
| 115 g | Na2HPO4 | |||
| 22.8 g | NaH2PO4 | |||
| Ultrapure water to 1 L | ||||
| Filter (0.22 μm) into a glass bottle. | ||||
| Add 0.1% DEPC and follow the instruction for DEPC water. | ||||
| 4X fixing solution (16% formaldehyde) | ||||
| 5 mL | 20% formaldehyde | |||
| 500 µL | DEPC water | |||
| 750 µL | 1M DEPC sodium phosphate buffer, pH 7.4 | |||
| Store at 4 °C for up to 2-4 weeks. | ||||
| CAUTION: Formaldehyde is toxic. Wear gloves and use a fume hood when making this solution. | ||||
| Wash solution | ||||
| Mix the following: | ||||
| 10 mL | Formamide (final 25%) | |||
| 4 mL | 20X SSC (final 2X) | |||
| Fill DEPC water to 40 mL | ||||
| Filter (0.22 μm) and store at 4 °C | ||||
| CAUTION: Formamide is toxic. Wear gloves and use a fume hood when making this solution. | ||||
| Pre-hybridization solution | ||||
| 200 µL | Formamide (final 20%) | |||
| 100 µL | 20X SSC (final 2X) | |||
| 10 µL | 100X VRC (final 1X) | |||
| 25 µL | 4% (w/v) BSA (final 0.1%) | |||
| 685 µL | DEPC water | |||
| NOTE: Vortex the VRC stock before taking 10 µL out. | ||||
| CAUTION: Formamide is toxic and a known teratogen. Wear gloves and handle it under a fume hood. | ||||
| Probe hybridization solution | ||||
| 200 µL | Formamide (final 20%) | |||
| 100 µL | 20X SSC (final 2X) | |||
| 10 µL | 100X VRC (final 1X) | |||
| 25 µL | 4% (w/v) BSA (final 0.1%) | |||
| 10 µL | 40 mg/mL E. coli tRNA (final 0.4 mg/mL) | |||
| 200 µL | 50% dextran sulfate (final 10%) | |||
| x µL | 5’ mRNA probe set (from Step 1.12) to final 4 nM. | |||
| y µL | 3’ mRNA probe set (from Step 1.12) to final 4 nM. | |||
| - | DEPC water to make the total volume 1 mL | |||
| NOTE: Add dextran sulfate last. Because it is very viscous, cut the end of a pipette tip before taking 200 μL out from the 50% stock. After adding dextran sulfate, pipette up and down to homogenize the solution. | ||||
Table 1: Recipes of the solutions used.
Discussion
Here, we presented a smFISH protocol for measuring mRNA kinetics in E. coli. In the previously published smFISH protocols for bacteria23, cells were kept in the tubes until the very end of the protocol, that is until they are ready for imaging. While it has many benefits, such as minimal nonspecific binding of fluorescent probes on the coverslip surface23, it is difficult to follow these protocols when there are many samples from a time-course experiment. First, a relatively large volume of cells (>1 mL) needs to be sampled and even harvested before fixation. Second, the cell samples need to be centrifuged multiple times to exchange solutions and to wash after the hybridization step. In our protocol, a small volume (<1 mL) of culture is directly mixed with a fixing solution in a 1.5-mL tube, helping to quickly “freeze” the cell state at the moment of sampling. Also, cells stay attached to the surface throughout the procedure, and different solutions can be exchanged quickly by aspirating liquids with a vacuum filtration system and applying solution drops at once with a multi-channel pipette. This difference makes our protocol highly advantageous when a large number of samples need to be processed at once. Using our protocol, 12-48 samples can be handled simultaneously and the entire FISH procedure can be completed within ~8 hours, about a similar amount of time needed for a few samples (Figure 2). Although we used the expression of lacZ in E. coli as an example, the protocol is widely applicable to different genes and bacterial species with considerations discussed below.
For different genes, the first thing to consider is smFISH probes. One may design oligonucleotide probes that tile the mRNA of interest (Figure 1A)13. In this “tiling” probe approach, each probe is ~20 base long and labeled with a fluorophore at the 5’ or 3’ terminus. This strategy is convenient as no genetic manipulation is needed. Alternatively, a tandem repeat of ~20 bp sequence, foreign to the genomic sequence (e.g., an array of lacO sequence in Caulobacter crescentus14), may be inserted in the untranslated region of a gene of interest and a single probe complementary to the repeat unit is used to label the mRNA (“array” approach; Figure 1B). In both cases, multiple fluorophores decorate an mRNA, giving amplified fluorescence signal that can be easily differentiated from a single probe nonspecifically bound inside a cell.
Whether to choose “tiling” or “array” approaches depends on the negative control, a sample where nonspecific binding of probes is tested because it lacks the target mRNA. For tiling probes (Figure 1A), a mutant strain without the gene of interest or a condition, in which the gene is not transcribed (e.g., repression of lacZ) can serve as a negative control for testing nonspecific binding of probes. For the array-based smFISH (Figure 1B), a wild-type strain lacking the array can serve as a negative control because it does not contain binding sites for the probes.
Optimal hybridization conditions may depend on the probe sequences and even the choice of fluorophore dyes. We optimized the hybridization condition for lacZ probe sets by keeping the hybridization temperature at 37 °C and testing different concentrations of probe sets and formamide in the hybridization solution. Higher concentrations of formamide tend to reduce both nonspecific and specific binding26,35. We recommend systematically changing the hybridization and its wash conditions while keeping hybridization time and temperature the same. As the condition becomes more stringent, both nonspecific and specific binding decrease (Figure 6). It is important to find a point where the nonspecific binding starts to hit below an acceptable threshold without further compromising specific binding. For example, we used the signal level obtained without any probes (“no probes”) as a threshold (Figure 6).
The two-color smFISH method labeling two separate regions of an mRNA is limited to long genes. To measure the rate of transcription elongation, we took advantage of the fact that lacZ is long (3075 bp) and its expression can be induced by IPTG. When a gene is short, it is difficult to design two tiling probe sets (near 5’ and 3’ ends) and resolve the time delay between appearances of 5’ vs. 3’ mRNA regions. In this case, one may count nascent mRNAs at steady state by smFISH and analyze their distribution with an analytical model that has the rate of transcription elongation as a fitting parameter20. Also, when a gene of interest is not inducible, one may treat cells with rifampicin at time zero and measure the temporal change in 5’ and 3’ mRNA sub-regions. The delay from the decrease of 5’ mRNA signal to that of 3’ mRNA signal can then be used to calculate the rate of transcription elongation as done previously31.
Finally, the smFISH protocol is versatile and can be combined with other labeling schemes. Previously, DNA locus was visualized together with mRNAs by combining mRNA FISH with either DNA FISH14 or fluorescent reporter-operator system20. Protein products may be visualized by performing immunofluorescence together with mRNA FISH14,36. Also, it can be combined with three-dimensional super-resolution microscopy37 to visualize mRNAs in all three dimensions38,39.
Disclosures
The authors declare they have no competing financial interests.
Acknowledgements
This protocol was developed by S.K. during her postdoctoral research in Dr. Christine Jacobs-Wagner's laboratory at the Howard Hughes Medical Institute and the Microbial Sciences Institute at Yale University. We thank Dr. Jacobs-Wagner and her lab members for various inputs during the method development and Laura Troyer for critical reading of the manuscript. S.K. acknowledges support from the Searle Scholars Program; K.V. acknowledges the support of James Scholar Preble Research Award from the University of Illinois.
Materials
| Name | Company | Catalog Number | Comments |
| Bacterial strain | |||
| Escherichia coli MG1655 | |||
| Chemicals, peptides, and others | |||
| Acetonitrile | Sigma-Aldrich | 34851 | UPLC buffer B |
| Ammonium chloride | Fisher Chemical | A661-500 | To make M9 medium |
| Bovine serum albumin (BSA) | Sigma-Aldrich | B2518 | Probe hybridization |
| Calcium chloride | Acros Organics | 349610250 | To make M9 medium |
| Casamino acid | BD Difco | 223050 | To make M9 medium |
| Cy3B NHS ester | GE Healthcare Life Sciences | PA63101 | Fluorophore for FISH probes |
| Cy5 NHS ester | GE Healthcare Life Sciences | PA15101 | Fluorophore for FISH probes |
| DEPC | Sigma-Aldrich | D5758 | |
| Dextran sulfate | Millipore | S4030 | Probe hybridization |
| Dextrose | Fisher Chemical | D16 | To repress the expression of lacZ |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D8418 | To dissolve fluorophores |
| E. coli tRNA | Sigma-Aldrich | R1753 | Probe hybridization |
| Ethanol | Decon Laboratories | 2701 | Used in DNA purification, lysis, and cleaning coverslips |
| FISH probes | Biosearch Technologies | Sequences are published in ref#16 | |
| Formaldehyde | Ladd Research Industries | 20295 | Fixation |
| Formamide | American Bio | AB00600 | Probe hybridization, pre-hybridization, and wash |
| Glycerol | Americanbio | AB00751-01000 | To make M9 medium |
| Isopropylthio-β-galactoside (IPTG) | Invitrogen | 15529019 | lacZ induction |
| Magnesium sulfate | Fisher Chemical | M65-500 | To make M9 medium |
| 2-Nitrophenyl β-D-fucopyranoside (ONPF) | Santa Cruz Biotechnology | sc-216258 | lacZ repression |
| Picodent twinsil 22 | Picodent | 1300 1000 | Sealant |
| Poly-L-lysine | Sigma-Aldrich | P8920 | To treat the coverslip surface |
| Potassium chloride | Fisher BioReagents | BP366-500 | To make PBS |
| Potassium phosphate monobasic | Fisher BioReagents | BP362-500 | To make PBS and M9 medium |
| Rifampicin | Sigma-Aldrich | R3501 | To stop transcription initiation |
| Saline-sodium citrate buffer (SSC) | Invitrogen | AM9763 | Probe hybridization, pre-hybridization, and wash |
| sodium bicarbonate | Fisher BioReagents | BP328-500 | Fluorophore-probe conjugation |
| Sodium chloride | Fisher BioReagents | BP358-1 | For DNA purification, PBS and M9 medium |
| Sodium phosphate dibasic | Fisher BioReagents | BP332-500 | To make PBS and M9 medium |
| Sodium phosphate monobasic | Fisher BioReagents | BP329-500 | To make a sodium phosphate buffer |
| Super PAP Pen | Invitrogen | 8899 | Hydrophobic marker for coverslips |
| TetraSpek microspheres | Invitrogen | T7280 | Controls for multi-channel registration |
| Thiamine | Sigma-Aldrich | T1270 | To make M9 medium |
| Triethylammonium acetate | Sigma-Aldrich | 90358 | UPLC buffer A |
| Vanadyl ribonucleoside complex (VRC) | Sigma-Aldrich | 94742 | Probe hybridization and pre-hybridization |
| Equipment | |||
| C18 column | Waters | Acquity BEH C18 column | |
| Countertop centrifuge | Eppendorf | 5425 | |
| Countertop incubator | Eppendorf | Thermomixer F1.5 | |
| Incubator (Oven) | Thermo Scientific | 51030514 | Gravity convection |
| Water purification system | Millipore | Milli-Q Reference | |
| Nanodrop | Thermo Scientific | 2000C | |
| Nitrogen gas | Building | For blow-drying coverslips and glass slides | |
| UPLC | Waters | Acquity UPLC system | |
| Vacuum and aspirator | Building | Aspirator is made of a filtration flask with a side arm. | |
| Vacuum concentrator | Labconco | 7810010 | Centrivap; to dry samples collected from UPLC. |
| Vortexer | Scientific Industries | Genie-2 SI-0236 | |
| Water bath shaker | New Brunswick | Innova 3100 | Critical for time-course experiments |
| Water bath sonicator | VWR | 97043-960 | To clean coverslips and glass slides |
| Tools | |||
| 1.5-mL tubes | Eppendorf | 22431021 | DNA lobind tubes |
| 1000-uL pipette tip box | Denville Scientific | P1126 | An empty box after using all the tips |
| Coplin jar | SPI | 01240-AB | To clean coverslips and glass slides |
| Coverslip | Fisher Scientific | 22-050-230 | 24x60 No1 |
| Filtered pipette tips | Denville Scientific | P1121,P1122,P1126 | SHARP® Precision Barrier Tips |
| Forceps | SPI | K35a | To handle clean coverslips and glass slides |
| Glass slide | Fisher Scientific | 12-544-1 | |
| Gloves | Microflex | MK-296-M | |
| Multichannel pipetter | Eppendorf | 2231300045 | To use in the washing step (#7) |
| Pipette | Gilson | P1000, P200, P20 | |
| Reagent reservoir | MTC Bio | P8025-1S | To use in the washing step (#7) |
| Syringe filter (0.22 um) | Millipore | SLGS033SS | |
| Timer | VWR | 62344-641 | |
| Software and algorithms | |||
| MATLAB | Mathworks | R2013 and up | https://www.mathworks.com |
| MicrobeTracker or Oufti | https://www.github.com/JacobsWagnerLab/MicrobeTracker | ||
| https://oufti.org/ | |||
| Stellaris Probe Designer | Biosearch Technologies | https://www.biosearchtech.com/support/tools/design-software/stellaris-probe-designer | |
| Microscope | |||
| CCD camera | Hamamatsu Photonics | Orca-II-ER | |
| Cy3 filter set | Chroma | 49004 | |
| Cy5 filter set | Chroma | 49006 | |
| Epi-fluorescence microscope | Nikon | Eclipse Ti | For phase-contrast and epi fluorescence |
| Fluorescence excitation source | Lumencor | SOLA-E | |
| Nikon Elements software | Nikon | software that controls the microscope setup | |
| Phase-contrast 100x objective | Nikon | Plan Apochromat (NA 1.45) | |
| Probe sequence | |||
| DNA oligos with C6 amino modification at the 5' end | Biosearch Technologies Inc | ||
| lacZ1 | GTGAATCCGTAATCATGGTC | 5' mRNA | |
| lacZ2 | TCACGACGTTGTAAAACGAC | 5' mRNA | |
| lacZ3 | ATTAAGTTGGGTAACGCCAG | 5' mRNA | |
| lacZ4 | TATTACGCCAGCTGGCGAAA | 5' mRNA | |
| lacZ5 | ATTCAGGCTGCGCAACTGTT | 5' mRNA | |
| lacZ6 | AAACCAGGCAAAGCGCCATT | 5' mRNA | |
| lacZ7 | AGTATCGGCCTCAGGAAGAT | 5' mRNA | |
| lacZ8 | AACCGTGCATCTGCCAGTTT | 5' mRNA | |
| lacZ9 | TAGGTCACGTTGGTGTAGAT | 5' mRNA | |
| lacZ10 | AATGTGAGCGAGTAACAACC | 5' mRNA | |
| lacZ11 | GTAGCCAGCTTTCATCAACA | 5' mRNA | |
| lacZ12 | AATAATTCGCGTCTGGCCTT | 5' mRNA | |
| lacZ13 | AGATGAAACGCCGAGTTAAC | 5' mRNA | |
| lacZ14 | AATTCAGACGGCAAACGACT | 5' mRNA | |
| lacZ15 | TTTCTCCGGCGCGTAAAAAT | 5' mRNA | |
| lacZ16 | ATCTTCCAGATAACTGCCGT | 5' mRNA | |
| lacZ17 | AACGAGACGTCACGGAAAAT | 5' mRNA | |
| lacZ18 | GCTGATTTGTGTAGTCGGTT | 5' mRNA | |
| lacZ19 | TTAAAGCGAGTGGCAACATG | 5' mRNA | |
| lacZ20 | AACTGTTACCCGTAGGTAGT | 5' mRNA | |
| lacZ21 | ATAATTTCACCGCCGAAAGG | 5' mRNA | |
| lacZ22 | TTTCGACGTTCAGACGTAGT | 5' mRNA | |
| lacZ23 | ATAGAGATTCGGGATTTCGG | 5' mRNA | |
| lacZ24 | TTCTGCTTCAATCAGCGTGC | 5' mRNA | |
| lacZ25 | ACCATTTTCAATCCGCACCT | ||
| lacZ26 | TTAACGCCTCGAATCAGCAA | ||
| lacZ27 | ATGCAGAGGATGATGCTCGT | ||
| lacZ28 | TCTGCTCATCCATGACCTGA | ||
| lacZ29 | TTCATCAGCAGGATATCCTG | ||
| lacZ30 | CACGGCGTTAAAGTTGTTCT | ||
| lacZ31 | TGGTTCGGATAATGCGAACA | ||
| lacZ32 | TTCATCCACCACATACAGGC | ||
| lacZ33 | TGCCGTGGGTTTCAATATTG | ||
| lacZ34 | ATCGGTCAGACGATTCATTG | ||
| lacZ35 | TGATCACACTCGGGTGATTA | ||
| lacZ36 | ATACAGCGCGTCGTGATTAG | ||
| lacZ37 | GATCGACAGATTTGATCCAG | ||
| lacZ38 | AAATAATATCGGTGGCCGTG | ||
| lacZ39 | TTTGATGGACCATTTCGGCA | ||
| lacZ40 | TATTCGCAAAGGATCAGCGG | ||
| lacZ41 | AAGACTGTTACCCATCGCGT | ||
| lacZ42 | TGCCAGTATTTAGCGAAACC | ||
| lacZ43 | AAACGGGGATACTGACGAAA | ||
| lacZ44 | TAATCAGCGACTGATCCACC | ||
| lacZ45 | GGGTTGCCGTTTTCATCATA | ||
| lacZ46 | TCGGCGTATCGCCAAAATCA | ||
| lacZ47 | TTCATACAGAACTGGCGATC | ||
| lacZ48 | TGGTGTTTTGCTTCCGTCAG | ||
| lacZ49 | ACGGAACTGGAAAAACTGCT | 3' mRNA | |
| lacZ50 | TATTCGCTGGTCACTTCGAT | 3' mRNA | |
| lacZ51 | GTTATCGCTATGACGGAACA | 3' mRNA | |
| lacZ52 | TTTACCTTGTGGAGCGACAT | 3' mRNA | |
| lacZ53 | GTTCAGGCAGTTCAATCAAC | 3' mRNA | |
| lacZ54 | TTGCACTACGCGTACTGTGA | 3' mRNA | |
| lacZ55 | AGCGTCACACTGAGGTTTTC | 3' mRNA | |
| lacZ56 | ATTTCGCTGGTGGTCAGATG | 3' mRNA | |
| lacZ57 | ACCCAGCTCGATGCAAAAAT | 3' mRNA | |
| lacZ58 | CGGTTAAATTGCCAACGCTT | 3' mRNA | |
| lacZ59 | CTGTGAAAGAAAGCCTGACT | 3' mRNA | |
| lacZ60 | GGCGTCAGCAGTTGTTTTTT | 3' mRNA | |
| lacZ61 | TACGCCAATGTCGTTATCCA | 3' mRNA | |
| lacZ62 | TAAGGTTTTCCCCTGATGCT | 3' mRNA | |
| lacZ63 | ATCAATCCGGTAGGTTTTCC | 3' mRNA | |
| lacZ64 | GTAATCGCCATTTGACCACT | 3' mRNA | |
| lacZ65 | AGTTTTCTTGCGGCCCTAAT | 3' mRNA | |
| lacZ66 | ATGTCTGACAATGGCAGATC | 3' mRNA | |
| lacZ67 | ATAATTCAATTCGCGCGTCC | 3' mRNA | |
| lacZ68 | TGATGTTGAACTGGAAGTCG | 3' mRNA | |
| lacZ69 | TCAGTTGCTGTTGACTGTAG | 3' mRNA | |
| lacZ70 | ATTCAGCCATGTGCCTTCTT | 3' mRNA | |
| lacZ71 | AATCCCCATATGGAAACCGT | 3' mRNA | |
| lacZ72 | AGACCAACTGGTAATGGTAG | 3' mRNA | |
References
- Bervoets, I., Charlier, D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiology Reviews. 43 (3), 304-339 (2019).
- Epshtein, V., Nudler, E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 300 (5620), 801-805 (2003).
- Vogel, U., Jensen, K. F. The RNA chain elongation rate in Escherichia coli depends on the growth rate. Journal of Bacteriology. 176 (10), 2807-2813 (1994).
- Tennyson, C. N., Klamut, H. J., Worton, R. G. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nature Genetics. 9, 184 (1995).
- Singh, J., Padgett, R. A. Rates of in situ transcription and splicing in large human genes. Nature Structural & Molecular Biology. 16, 1128 (2009).
- Selinger, D. W., Saxena, R. M., Cheung, K. J., Church, G. M., Rosenow, C. Global RNA Half-Life Analysis in Escherichia coli Reveals Positional Patterns of Transcript Degradation. Genome Research. 13 (2), 216-223 (2003).
- Bernstein, J. A., Khodursky, A. B., Lin, P. -. H., Lin-Chao, S., Cohen, S. N. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proceedings of the National Academy of Sciences. 99 (15), 9697-9702 (2002).
- Pérez-Ortín, J. E., Medina, D. A., Chávez, S., Moreno, J. What do you mean by transcription rate. BioEssays. 35 (12), 1056-1062 (2013).
- Tang, F., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nature Methods. 6 (5), 377-382 (2009).
- Kuchina, A., et al. Microbial single-cell RNA sequencing by split-pool barcoding. BioRxiv. , 869248 (2019).
- Blattman, S. B., Jiang, W., Oikonomou, P., Tavazoie, S. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nature Microbiology. , (2020).
- Femino, A., Fay, F., Fogarty, K., Singer, R. Visualization of single RNA transcripts in situ. Science. 280 (5363), 585-590 (1998).
- Raj, A., vanden Bogaard, P., Rifkin, S. A., van Oudenaarden, A., Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 5 (10), 877-879 (2008).
- Montero Llopis, P., et al. Spatial organization of the flow of genetic information in bacteria. Nature. 466 (7302), 77-81 (2010).
- So, L. -. h., et al. General properties of transcriptional time series in Escherichia coli. Nature Genetics. 43 (6), 554-560 (2011).
- Taniguchi, Y., et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 329 (5991), 533-538 (2010).
- Jones, D. L., Brewster, R. C., Phillips, R. Promoter architecture dictates cell-to-cell variability in gene expression. Science. 346 (6216), 1533-1536 (2014).
- Iyer, S., Park, B. R., Kim, M. Absolute quantitative measurement of transcriptional kinetic parameters in vivo. Nucleic Acids Research. 44 (18), 142 (2016).
- Kim, S., Beltran, B., Irnov, I., Jacobs-Wagner, C. Long-Distance Cooperative and Antagonistic RNA Polymerase Dynamics via DNA Supercoiling. Cell. 179 (1), 106-119 (2019).
- Wang, M., Zhang, J., Xu, H., Golding, I. Measuring transcription at a single gene copy reveals hidden drivers of bacterial individuality. Nature Microbiology. 4 (12), 2118-2127 (2019).
- Joo, C., Ha, T. . Labeling DNA (or RNA) for single-molecule FRET. 2012 (9), 1005-1008 (2012).
- Sambrook, J., Russell, D. W. . Standard Ethanol Precipitation of DNA in Microcentrifuge Tubes. 2006 (1), 4456 (2006).
- Skinner, S. O., Sepúlveda, L. A., Xu, H., Golding, I. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nature Protocols. 8 (6), 1100-1113 (2013).
- Adesnik, M., Levinthal, C. The synthesis and degradation of lactose operon messenger RNA in E. coli. Cold Spring Harbor Symposia on Quantitative Biology. 35, 451-459 (1970).
- Campbell, E. A., et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 104 (6), 901-912 (2001).
- Raj, A., Tyagi, S., Walter, N. G. . Methods in Enzymology. 472, 365-386 (2010).
- Sliusarenko, O., Heinritz, J., Emonet, T., Jacobs-Wagner, C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Molecular Microbiology. 80 (3), 612-627 (2011).
- Paintdakhi, A., et al. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Molecular Microbiology. 99 (4), 767-777 (2016).
- Moffitt, J. R., Zhuang, X., Filonov, G. S., Jaffrey, S. R. . Methods in Enzymology. 572, 1-49 (2016).
- Yu, J., Xiao, J., Ren, X., Lao, K., Xie, X. S. Probing gene expression in live cells, one protein molecule at a time. Science. 311 (5767), 1600-1603 (2006).
- Chen, H., Shiroguchi, K., Ge, H., Xie, X. S. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Molecular Systems Biology. 11 (1), 781 (2015).
- Vogel, U., Sørensen, M., Pedersen, S., Jensen, K. F., Kilstrup, M. Decreasing transcription elongation rate in Escherichia Coli exposed to amino acid starvation. Molecular Microbiology. 6 (15), 2191-2200 (1992).
- Yang, S., et al. Transcription and translation contribute to gene locus relocation to the nucleoid periphery in E. coli. Nature Communications. 10 (1), 5131 (2019).
- Zenklusen, D., Larson, D. R., Singer, R. H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature Structural & Molecular Biology. 15 (12), 1263-1271 (2008).
- Fontenete, S., Guimarães, N., Wengel, J., Azevedo, N. F. Prediction of melting temperatures in fluorescence in situ hybridization (FISH) procedures using thermodynamic models. Critical Reviews in Biotechnology. 36 (3), 566-577 (2016).
- Sepúlveda, L. A., Xu, H., Zhang, J., Wang, M., Golding, I. Measurement of gene regulation in individual cells reveals rapid switching between promoter states. Science. 351 (6278), 1218-1222 (2016).
- Huang, B., Wang, W., Bates, M., Zhuang, X. Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science. 319 (5864), 810-813 (2008).
- Moffitt, J. R., Pandey, S., Boettiger, A. N., Wang, S., Zhuang, X. Spatial organization shapes the turnover of a bacterial transcriptome. eLife. 5, 13065 (2016).
- Fei, J., et al. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science. 347 (6228), 1371-1374 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved