A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Cell Dissociation from the Tongue Epithelium and Mesenchyme/Connective Tissue of Embryonic-Day 12.5 and 8-Week-Old Mice
In This Article
Summary
We have developed a generalized protocol to dissociate a large quantity of high-quality single cells from the epithelium and mesenchyme/connective tissue of embryonic and adult mouse tongues.
Abstract
Cell dissociation has been an essential procedure for studies at the individual-cell level and/or at a cell-population level (e.g., single cell RNA sequencing and primary cell culture). Yielding viable, healthy cells in large quantities is critical, and the optimal conditions to do so are tissue dependent. Cell populations in the tongue epithelium and underlying mesenchyme/connective tissue are heterogeneous and tissue structures vary in different regions and at different developmental stages. We have tested protocols for isolating cells from the mouse tongue epithelium and mesenchyme/connective tissue in the early developmental [embryonic day 12.5 (E12.5)] and young adult (8-week) stages. A clean separation between the epithelium and underlying mesenchyme/connective tissue was easy to accomplish. However, to further process and isolate cells, yielding viable healthy cells in large quantities, and careful selection of enzymatic digestion buffer, incubation time, and centrifugation speed and time are critical. Incubation of separated epithelium or underlying mesenchyme/connective tissue in 0.25% Trypsin-EDTA for 30 min at 37 °C, followed by centrifugation at 200 x g for 8 min resulted in a high yield of cells at a high viability rate (>90%) regardless of the mouse stages and tongue regions. Moreover, we found that both dissociated epithelial and mesenchymal/connective tissue cells from embryonic and adult tongues could survive in the cell culture-based medium for at least 3 h without a significant decrease of cell viability. The protocols will be useful for studies that require the preparation of isolated cells from mouse tongues at early developmental (E12.5) and young adult (8-week) stages requiring cell dissociation from different tissue compartments.
Introduction
The mammalian tongue is a complex organ critical for taste, speaking, and food processing. It is comprised of multiple types of highly organized tissues compartmentalized by mesenchyme/connective tissue and covered by a stratified epithelial sheet containing taste papillae and taste buds. Cell populations in both tongue epithelium and mesenchyme/connective tissue are heterogeneous. To better understand the functions and distribution of a particular type of cells in the tongue, studies using dissociated cells are necessary. For example, single cell RNA sequencing is a powerful and high-throughput method for transcriptomic profiling in individual cells, which is designed to understand the transcriptome of complex tissue at a single-cell resolution1,2,3,4. Primary cell culture has been proven to be a useful tool to study the function and differentiation of stem/progenitor cells for taste buds5,6. These studies require a large quantity of high-quality isolated cell populations (e.g., sufficient total cell number with proper concentration and high viability).
Thus, there is a need to isolate cells from different regions of the lingual tissues and at different developmental stages. Currently, there is not a detailed protocol available for cell dissociation from the tongue epithelium and underlying mesenchyme/connective tissue. Here, we report an optimized cell dissociation method to prepare cells for experiments requiring a high quality of live cells such as for single cell RNA sequencing and primary stem cell cultures. We found that selection of enzymatic digestion buffer, gentle pipetting, selection of resuspension medium, and optimal centrifugation time and speed are crucial to generate these large quantities of high-quality cells.
Protocol
Animal use (C57BL/6 mice throughout the study) was approved by the University of Georgia Institutional Animal Care and Use Committee and was in accordance with the National Institutes of Health Guidelines for care and use of animals for research.
1. Animal usage
NOTE: Mice were bred and maintained in the animal facility of the Animal and Dairy Science department at the University of Georgia at 22 °C under 12-h day/night cycles.
- Designate noon of the day of vaginal plug detection in mice as embryonic (E) day 0.5. Use embryos at E12.5 and postnatal mice at 8 weeks of age for the following experiments.
2. Preparation before experiment
NOTE: The instruments required for this protocol are listed in the Table of Materials.
- Autoclave instruments before the experiment. Sterilize instruments using a bead sterilizer during the experiment.
- Clean the surgical area, the dissecting microscope, and the biosafety cabinet using 70% ethanol wipes. Turn on the UV light of biosafety cabinet and keep it on for 20 min prior to the experimental procedure.
- Prepare an enzyme mixture of 1:1 dispase (5.0 mg/mL) and collagenase (2.0 mg/mL) to a final concentration of 2.5 and 1.0 mg/mL respectively, and filter the solution using 0.22 µm syringe filter7.
- Prepare 1 mL of enzyme mixture for an adult tongue or 0.5 mL for a specific region of tongue (e.g., posterior, or anterior tongue).
- Prepare 2 mL of enzyme mixture for embryonic tongues.
- Make 10 mL of 2.5% BSA in 0.1 M PBS and filter the solution using 1 mL syringe and 0.22 µm syringe filter.
- Make 500 µL of 5% FBS in DMEM/F12.
- Make 3 mL of DMEM/F12 containing 10% FBS and 1% BSA and filter the solution using 1 mL syringe and 0.22 µm syringe filter.
3. Separation of the tongue epithelium from the mesenchyme/underlying connective tissue
- Separation of the epithelium from the mesenchyme of an E12.5 mouse tongue
- Euthanize timed pregnant female mice carrying E12.5 embryos by placing it in a CO2 chamber followed by cervical dislocation.
NOTE: Mouse embryos at E12.5 are collected after 12 pm (afternoon) on the 12th day after detection of vaginal plug in the pregnant female mice. - Transfer mice to the surgical area. Wet the mouse abdomen using 70% ethanol to prevent fur from getting into the operating site.
- Open the abdomen using dissecting scissors to expose the uterine horns carrying embryos. Dissect the uterine horns using dissecting scissors and transfer it to 15 mL of fresh Tyrode's solution in a 100 mm culture dish.
- Dissect the embryos (Figure 1A1) out from uterine horns under a dissecting microscope using mini-scissors and fine forceps.
- Carefully open the mouth cavity wide by using fine forceps and dissect the tongue off from the mandible using mini-scissors (Figure 1A2).
- Wash the tongues using 15 mL of fresh sterile Tyrode's solution in a 100 mm culture dish.
- Transfer the tissues to 2 mL of enzyme mixture of dispase (2.5 mg/mL) and collagenase (1.0 mg/mL) in a 35 mm culture dish with a spatula and fine forceps in a biosafety cabinet. Incubate for 20 min at 37 °C.
- Transfer tongues to 15 mL of fresh sterile Tyrode's solution in a 100 mm culture dish and gently remove the mesenchyme from the epithelium from the ventral side using fine forceps.
NOTE: Epithelial sheets may be separated without mechanical force during the incubation. - Wash the separated epithelia and mesenchyme twice in 15 mL of fresh sterile Tyrode's solution in a 100 mm culture dish.
NOTE: The activities of dispase and collagenase will be inhibited by EDTA in the cell dissociation procedure (step 4.1).
- Euthanize timed pregnant female mice carrying E12.5 embryos by placing it in a CO2 chamber followed by cervical dislocation.
- Separation of the tongue epithelium from the underlying connective tissue of adult mice
- Euthanize the mouse at 8 weeks of age by placing it in a CO2 chamber. Confirm that the mouse is euthanized with no breaths and forepaw-pinch response.
- Transfer the mice to the surgical area. Wet the mouse head using 70% ethanol to prevent fur from getting into the oral cavity.
- Cut the corners of the mouth along the cheek using dissecting scissors to open the oral cavity. Dissect the tongue with mandible (Figure 1B1) and place it in a plastic dish with a layer of plastic wrap.
- Using surgical forceps to hold the tongue under a dissecting microscope, inject the enzyme mixture of dispase (2.5 mg/mL) and collagenase (1.0 mg/mL) in the sub-epithelial space of tongue through the cutting edge (Figure 1B1, arrows) of the posterior tongue.
- Inject 1 mL of enzyme mixture evenly to the whole tongue for tissue collection from both anterior and posterior tongue.
- Inject 0.5 mL of enzyme mixture locally to the anterior tongue for tissue collection from tongue tip or to the posterior tongue for circumvallate papilla tissue collection.
NOTE: The tongue will swell as the enzyme accumulates (Figure 1B2). Gentle injection of the enzyme can prevent pressure from damaging the epithelium and keep as much enzyme as possible in the tongue.
- Wrap the tongue with plastic wrap and incubate the tongue for 30 min at 37 °C.
- Use mini-scissors to dissect the tongue tip and/or circumvallate papilla, and use spatula and fine forceps to transfer tissue to 15 mL of fresh sterile Tyrode's solution in a 100 mm culture dish.
- Separate the epithelium from the underlying connective tissue in the enzyme-digested sub-epithelial space using mini-scissors. Trim the tissues to a proper size according to the requirement of downstream experiments.
- Wash the separated epithelium and underlying connective tissue twice in 15 mL of fresh sterile Tyrode's solution in a 100 mm culture dish.
NOTE: The activities of dispase and collagenase will be inhibited by EDTA in the cell dissociation procedure (step 4.1).
4. Cell dissociation
NOTE: The cell dissociation protocol described here can be applied to the tongue epithelium and mesenchyme/connective tissue in both E12.5 embryonic and 8-week-old mice. To reduce the cell loss during agitation and transfer of cell suspension, use commercial low retention pipette tips or pre-coated pipette tips with 2.5% BSA in 0.1 M PBS at pH 7.48.
- Transfer the tissues using spatula and fine forceps to 3 mL of 0.25% trypsin-EDTA in a new 35 mm culture dish for 30 min at 37 °C. Gently agitate tissues every 5 min with 1 mL pipette tips.
NOTE: Do not cut the pipette tip, as the cutting edge can physically damage the dissociated cells. - Add 500 µL of 5% FBS in DMEM/F12 to stop the reaction and transfer the medium to a 5 mL low binding centrifuge tube.
- Centrifuge cell suspension at 200 x g for 8 min at room temperature and remove the supernatant.
- Gently re-suspend cells in 3 mL of DMEM/F12 containing 10% FBS and 1% BSA using 1 mL pipette tips and filter the cells using a 70 µm cell strainer, followed by a 35 µm cell strainer.
- Centrifuge cell suspension at 200 x g for 8 min at room temperature. Remove most of the medium and leave 50-300 µL as the final volume to re-suspend cells.
NOTE: Adjust the concentration of cells according to requirements of downstream experiments by changing the final volume of single cell suspension.
5. Cell counting and viability test using hemocytometer
NOTE: To improve measurement accuracy, 3 technical replicates are recommended for each sample.
- Gently mix 5-10 µL of the single cell suspension with an equal volume of Trypan blue and add onto hemocytometer.
NOTE: Clean the hemocytometer thoroughly using 70% ethanol before use. Dusts particles on the hemocytometer will be stained in dark blue and affect the accuracy of viability test. Check the hemocytometer under a microscope. - Count the total cell number, the number of live cells (white), and the number of dead cells (dark blue) stained by Trypan blue (Figure 2, arrows) respectively in 4 squares with 16 grids (Figure 2) using inverted microscope with imaging system.
- Calculate the cell concentration:
Cell concentration =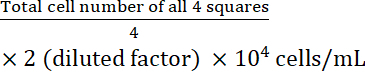
- Calculate the viability:
Viability =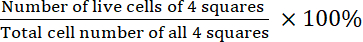
- Calculate the viability:
Results
Separation of the tongue epithelium from the underlying mesenchyme/connective tissue
In the embryonic mouse tongue, a gap in the sub-epithelial space is visible after proper enzyme digestion. Epithelial sheets of some tongues are separated without mechanical force during the incubation.
In the adult mouse tongue, a successful enzyme injection is indicated by the swelling in the injected areas (Figure 1B2), which s...
Discussion
To date, there has not been a detailed protocol available for cell dissociation from the tongue epithelium and underlying mesenchyme/connective tissue. This current cell dissociation protocol provides a reproducible procedure to generate a single cell suspension with a high cell viability (>90%) from mouse tongue tissues, including epithelial sheets and mesenchyme/connective tissues at both embryonic and postnatal stages even though isolated cells from E12.5 and adult mice are different in size. For example, isolated ...
Disclosures
No conflicts of interest declared.
Acknowledgements
This study was supported by the National Institutes of Health, grant number R01DC012308 and R21DC018089 to HXL. We give thanks to Brett Marshall (University of Georgia, Athens, GA) and Egon Ranghini (10X GENOMICS, Pleasanton, CA) for technical assistance and consultation regarding the cell dissociation; to Francisca Gibson Burnley (University of Georgia, Athens, GA) for English editing.
Materials
| Name | Company | Catalog Number | Comments |
| bovine serum albumin (BSA) | Gold Biotechnology | A-420-100 | |
| C57BL/6 mouse (C57BL/6J) | The Jackson Laboratory | 000664 | |
| collagenase (Collagenase A) | Sigma-Aldrich | 10103586001 | |
| culture dish (35 mm in diameter) | Genesee Scientific | 32-103G | |
| culture dish (100 mm in diameter) | Genesee Scientific | 32-107G | |
| dispase (Dispase II) | Sigma-Aldrich | 04942078001 | |
| dissecting scissors (Student Fine Scissors) | Find Science Tool | 91460-11 | |
| DMEM/F12 | Gibco | 11320033 | |
| fetal bovine serum (FBS) | Hyclone | C838U82 | |
| fine forceps (Dumount #3 Forceps) | Find Science Tool | 11293-00 | |
| hemocytometer | Hausser Scientific | 3520 | |
| inverted microscope with imaging system (EVOS XL Core Cell Imaging System) | Life Technologies | AMEX1000 | |
| low retention pipette tips | METTLER TOLEDO | 17014342 | |
| mini-scissors (Evo Spring Scissors) | Fine Science Tool | 15800-01 | |
| plastic warp | VWR | 46610-056 | |
| spatula (Moria Spoon) | Fine Science Tool | 10321-08 | |
| surgical forceps (Dumount #2 Laminectomy Forceps) | Fine Science Tool | 11223-20 | |
| Trypan blue | Gibco | 15250061 | |
| Tyrode’s solution | Sigma-Aldrich | T2145-10L | made from Tyrode's salts |
| 0.25% typsin-EDTA | Gibco | 25200056 | |
| 0.1 M Phosphate-Buffered Saline (PBS) | Hoefer | 33946 | made from 1 M PBS |
| 0.22-μm syringe filter | Genesee Scientific | 25-243 | |
| 70% ethanol | Koptec | 233919 | made from 100% ethanol |
| 1-mL syringe | BD | 8194938 | |
| 5-mL low binding microcentrifuge tube | Eppendorf | 30122348 | |
| 30-G needle | BD | 9193532 | |
| 35-μm cell strainer | Falcon | 64750 | |
| 70-μm cell strainer | Falcon | 64752 |
References
- Grada, A., Weinbrecht, K. Next-generation sequencing: methodology and application. The Journal of investigative dermatology. 133 (8), 11 (2013).
- Whitley, S. K., Horne, W. T., Kolls, J. K. Research techniques made simple: methodology and clinical applications of RNA sequencing. Journal of Investigative Dermatology. 136 (8), 77-82 (2016).
- Schaum, N., et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris: The Tabula Muris Consortium. Nature. 562 (7727), 367 (2018).
- Sukumaran, S. K., et al. Whole transcriptome profiling of taste bud cells. Scientific reports. 7 (1), 1-15 (2017).
- Ren, W., et al. Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation. Scientific Reports. 7 (1), 1-13 (2017).
- Ren, W., et al. Single Lgr5-or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proceedings of the National Academy of Sciences. 111 (46), 16401-16406 (2014).
- Venkatesan, N., Boggs, K., Liu, H. -. X. Taste bud labeling in whole tongue epithelial sheet in adult mice. Tissue Engineering Part C: Methods. 22 (4), 332-337 (2016).
- Nguyen-Ngoc, K. -. V., et al. . Tissue Morphogenesis. , 135-162 (2015).
- Okubo, T., Clark, C., Hogan, B. L. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells. 27 (2), 442-450 (2009).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved