Sign In
A subscription to JoVE is required to view this content. Sign in or start your free trial.
Methods Article
Optical Tweezers to Study RNA-Protein Interactions in Translation Regulation
In This Article
Summary
This protocol presents a complete experimental workflow for studying RNA-protein interactions using optical tweezers. Several possible experimental setups are outlined including the combination of optical tweezers with confocal microscopy.
Abstract
RNA adopts diverse structural folds, which are essential for its functions and thereby can impact diverse processes in the cell. In addition, the structure and function of an RNA can be modulated by various trans-acting factors, such as proteins, metabolites or other RNAs. Frameshifting RNA molecules, for instance, are regulatory RNAs located in coding regions, which direct translating ribosomes into an alternative open reading frame, and thereby act as gene switches. They may also adopt different folds after binding to proteins or other trans-factors. To dissect the role of RNA-binding proteins in translation and how they modulate RNA structure and stability, it is crucial to study the interplay and mechanical features of these RNA-protein complexes simultaneously. This work illustrates how to employ single-molecule-fluorescence-coupled optical tweezers to explore the conformational and thermodynamic landscape of RNA-protein complexes at a high resolution. As an example, the interaction of the SARS-CoV-2 programmed ribosomal frameshifting element with the trans-acting factor short isoform of zinc-finger antiviral protein is elaborated. In addition, fluorescence-labeled ribosomes were monitored using the confocal unit, which would ultimately enable the study of translation elongation. The fluorescence coupled OT assay can be widely applied to explore diverse RNA-protein complexes or trans-acting factors regulating translation and could facilitate studies of RNA-based gene regulation.
Introduction
Transfer of genetic information from DNA to proteins through mRNAs is a complex biochemical process, which is precisely regulated on all levels through macromolecular interactions inside cells. For translational regulation, RNA-protein interactions confer a critical role to rapidly react to various stimuli and signals1,2. Some RNA-protein interactions affect mRNA stability and thereby alter the time an RNA is translationally active. Other RNA-protein interactions are associated with recoding mechanisms such as stop-codon readthrough, bypassing, or programmed ribosomal frameshifting (PRF)3,4,5,6,7. Recently, a number of RNA-binding proteins (RBPs) have been demonstrated to interact with stimulatory mRNA elements and the translation machinery to dictate when and how much recoding will occur in the cell7,8,9,10,11. Thus, to dissect the role of RNA-binding proteins in translation and how they modulate RNA structure and stability, it is pivotal to study the interaction principles and mechanical properties of these RNA-protein complexes in detail.
Decades of work have laid the foundation to study the multi-step and multi-component process of translation, which relies on intricate communication between the RNA and protein components of the translation machinery to achieve speed and accuracy12,13,14. A crucial next step in understanding complex regulatory events is determining the forces, timescales, and structural determinants during translation at high precision12,15,16,17. The study of RNA conformational dynamics and especially how trans-acting auxiliary factors act on the RNA structure during translation have been further illuminated by the emergence of single-molecule tools, including optical tweezers or zero-mode waveguides16,17,18,19,20,21,22,23,24,25,26.
Optical tweezers (OT) represent a highly precise single-molecule technique, which has been applied to study many sorts of RNA-dependent dynamic processes including transcription, and translation26,27,28,29,30,31,32. The use of optical tweezers has allowed probing of molecular interactions, nucleic acid structures, and thermodynamic properties, kinetics, and energetics of these processes in detail16,17,22,33,34,35,36,37,38,39. Optical tweezers assay is based on the entrapment of microscopic objects with a focused laser beam. In a typical OT experiment, the molecule of interest is tethered between two transparent (usually polystyrene) beads (Figure 1A)27. These beads are then caught by optical traps, which behave like springs. Thus, the force applied on the molecule can be calculated based on the bead's displacement from the center of the focused laser beam (trap center). Recently, optical tweezers have been combined with confocal microscopy (Figure 1B), enabling fluorescence or Förster resonance energy transfer (FRET) measurements40,41,42. This opens a whole new field of possible experiments allowing simultaneous measurement and, therefore, precise correlation of force spectroscopy and fluorescence data.
Here, we demonstrate experiments using the optical tweezers combined with confocal microscopy to study protein-RNA interactions regulating translational frameshifting. Between the objective and the condenser, a flow cell with five channels enables continuous sample application with laminar flow. Through the microfluidic channels, various components can be injected directly, which decreases the hands-on time as well as allowing very little sample consumption throughout the experiment.
First, a basic guideline to assist the design of OT experiments is proposed and advantages as well as pitfalls of various setups are discussed. Next, the preparation of samples and experimental workflows are described, and a protocol for the data analysis is provided. To represent an example, we outline the results obtained from RNA stretching experiments to study the SARS-CoV-2 frameshifting RNA element (Figure 2A) with the trans-acting factor the short isoform of zinc-finger antiviral protein (ZAP), which alters the translation of the viral RNA from an alternative reading frame43. Additionally, it is demonstrated that fluorescence-labeled ribosomes can be employed in this OT confocal assay, which would be useful to monitor the processivity and speed of the translation machinery. The method presented here can be used to rapidly test the effect of different buffers, ligands, or other cellular components to study various aspects of translation. Finally, common experimental pitfalls and how to troubleshoot them are discussed. Below, some crucial points in experimental design are outlined.
Construct design
In principle, there are two common approaches to create an OT-compatible RNA construct. The first approach employs a long RNA molecule that is hybridized with complementary DNA handles, thus yielding a construct consisting of two RNA/DNA hybrid regions flanking a single-stranded RNA sequence in the middle (Figure 2B). This approach is employed in most OT RNA experiments33,44,45.
The second approach takes advantage of dsDNA handles with short (around 20 nt) overhangs15,17. These overhangs are then hybridized with the RNA molecule. Although more complicated in design, the use of dsDNA handles overcomes some of limitations of the DNA/RNA-hybrid system. In principle, even very long handles (>10kb) can be implemented, which is more convenient for confocal measurements. In addition, the RNA molecule can be ligated to DNA handles to increase tether stability.
End-labeling strategy
The construct must be tethered to beads via a strong molecular interaction. While there are approaches available for covalent bonding of handles to beads46, strong but non-covalent interactions such as streptavidin-biotin and digoxigenin-antibody are commonly used in OT experiments15,33,35,45. In the described protocol, the construct is labeled with biotin or digoxigenin, and the beads are coated with streptavidin or antibodies against digoxigenin, respectively (Figure 1A). This approach would be suitable for applying forces up to approximately 60 pN (per tether)47. Furthermore, the use of different 5' and 3' labeling strategies allow determining the orientation of the tether formed between the beads17.
Protein labeling for fluorescence measurements
For the confocal imaging, there are several commonly used approaches for fluorescence labeling. For instance, fluorophores can be covalently attached to amino acid residues that are found natively in proteins or introduced by site-directed mutagenesis through a reactive organic group. Thiol or amine-reactive dyes can be used for labeling of cysteine and lysine residues, respectively. There are several reversible protection methods to increase the specificity of labeling48,49, however native proteins would typically be labeled at multiple residues. Although the small size of the fluorophore may confer an advantage, non-specific labeling might interfere with the protein activity and thus signal intensity may vary49. Also, depending on the labeling efficiency signal intensity may differ between different experiments. Therefore, an activity check should be performed prior to the experiment.
In case the protein of interest contains an N- or C-terminal tag, such as a His-tag or strep-tag, specific labeling of these tags represents another popular approach. Moreover, tag-targeted labeling reduces the chance of the fluorophore interfering with protein activity and can enhance solubility49. However, tag-specific labeling usually yields mono-fluorophore labeled proteins, which might be challenging to detect. Another way of specific labeling can be accomplished by employing antibodies.
Microfluidics setup
The combination of OT with a microfluidics system allows a rapid transition between different experimental conditions. Moreover, current systems take advantage of maintaining the laminar flow inside the flow cell, which precludes the mixing of liquids from other channels in the perpendicular direction relative to the flow direction. Therefore, laminar flow is particularly advantageous for the experimental design. Currently, flow cells with up to 5 channels are commonly employed (Figure 3).
Protocol
1. Sample preparation
- Clone the sequence of interest into the vector containing the Lambda DNA fragments, which serve as the handle sequences (Figure 2)43,50.
- First generate a DNA template for subsequent in vitro transcription via PCR (Figure 2B; reaction 1). At this PCR step, the T7 promoter is added in the 5' end of the sense DNA molecule32,33,43,50. Set the PCR reaction according to Table 1. Run the PCR in 50 µL aliquots with appropriate cycles in the thermocycler.
- Prepare the handles by two separate PCR reactions (Table 1, Figure 2B; reaction 2 and 3). First, generate the 5' handle by PCR. Then, generate the 3' handle and simultaneously label it with digoxigenin by using a 5' digoxigenin-labeled primer32,33,43,50.
- After the PCR, purify the DNA using silica spin columns.
- Carry out the in vitro transcription reaction using T7 RNA polymerase (Table 2)32,33,43,50. Incubate the reaction at 37 °C for 2-4 h depending on the length of the RNA. Next, add DNase I to the reaction and incubate at 37 °C for 30 min to digest the DNA template. Purify the RNA using silica spin columns.
- During the labeling reaction of the 5' handle (Table 3), add biotin-16-dUTP at the 3' end of the handle by T4 DNA polymerase38,50. Perform the reaction at room temperature for 1-2 h. Afterwards, purify the DNA using silica spin columns.
NOTE: Since the 5' handle must be labeled at its 3' end (Figure 2B), the labeling cannot be performed during the PCR. - Mix the components mentioned above - 5' handle (3' labeled with biotin), 3' handle (5' labeled with digoxigenin), and RNA - in a 1:1:1 molar ratio in annealing buffer (80% formamide, 400 mM NaCl, 40 mM HEPES, pH 7.5, 0.5 mM EDTA, pH 8), to obtain the desired RNA/DNA hybrid (Table 4). Heat the annealing mixture up to 85 °C for 10 min and then slowly cool down to 4 °C.
- Mix the annealed sample with 1/10 of volume of 3 M sodium acetate (pH 5), 3 volumes of ice-cold ethanol and incubate at -80 °C for at least 1 h or at -20 °C overnight.
- Centrifuge the samples at 15,000 × g for 30 min at 4 °C. Discard the supernatant and dry the pellet (usually not visible) under vacuum.
- Finally, resuspend, the pellet in 50 µL of RNase-free water and make aliquots. Store the aliquots at -80 °C until used. For short term storage, the samples can be also stored at -20 °C.
2. Instrument setup
NOTE: The following protocol is optimized for the commercial optical tweezers instrument C-Trap from LUMICKS company. Therefore, adjustments to the presented steps might be necessary while using other optical tweezers instruments. If not used, the microfluidics system of the machine is kept in bleach (sodium hypochlorite solution) and must be washed before use.
- Discard the bleach and fill the syringes with 1 mL of RNase-free water.
- Add 50 µL of 0.5 M sodium thiosulfate to at least 1 mL of the RNase-free water and thoroughly wash the system (1 bar, at least 0.5 mL) to eliminate the remaining bleach in the system.
- Discard the sodium thiosulfate solution from the syringes. Replace syringes with fresh ones and wash the system with at least 0.5 mL of RNase-free water.
NOTE: Be careful, that the microfluidics system never runs dry to avoid air bubbles in the system. - Put 2 drops of immersion oil (refractive index of 1.33) or approximately 70 µL of water on top of the objective.
- Place the flow cell inside the holding frame in its position.
- Put 2 drops of immersion oil (refractive index of 1.51) on top of the flow cell.
- Turn on the laser device in the tweezers machine. Once it is running, turn on the trapping laser in the software interface at 100%.
- Using diagnostic cameras (Z-finder), adjust the Z-axis to the middle of the chamber between the second and the third reflections (interfaces) where the refraction rings are the biggest, by turning the micro screw.
NOTE: Each time the objective is moved closer to the measuring chamber and the focal plane of the objective crosses the interface between two phases, a reflection can be recognized in the Z-finder mode. There are 4 interfaces possible: (i) water/immersion oil and bottom glass (ii) bottom glass and buffer inside the chamber (iii) buffer inside the chamber and top glass (iv) top glass and immersion oil for condenser. - Adjust the condenser position (set trapping laser to approximately 50%) so the condenser touches the immersion oil on top of the measuring chamber.
- Adjust the focus by moving slowly down/up with the condenser, so approx. 10 light bands are shown in the moon mode (diagnostic cameras).
3. Sample measurement
- Incubate anti-digoxigenin-coated beads (AD) with the sample constructs (3 µL of 0.1% (w/v) AD bead suspension + 4 µL of sample) and with 1 µL of RNase inhibitors and 8 µL of the assay buffer (300 mM KCl, 5 mM MgCl2, 20 mM HEPES, pH 7.6, 0.05% Tween 20, 5 mM DTT) at RT for 10-20 min. After the incubation, dilute the sample in 500 µL of assay buffer.
NOTE: It is recommended to add oxygen scavengers, particularly during fluorescence measurements to the buffer in order to prevent oxidative damage. Here oxygen scavenger system containing glucose (8.3 mg/mL), glucose oxidase (40 U/mL) and catalase (185 U/mL) was used. - Mix 0.8 µL of 1% (w/v) streptavidin-coated (SA) beads with 1 mL of assay buffer.
- Discard water from the syringes and fill the syringes with respective suspensions/solutions. Wash for at least 2 min at approximately 1 bar, and then start catching beads.
NOTE: Depending on the experimental set-up, different channel arrangements may be used (Figure 3). Typically, one flow channel is filled with anti-digoxigenin beads carrying the RNA molecule. A second channel is filled with the streptavidin-coated beads. Buffer channel is used to form the tethers. A fourth channel can be employed to load the RNA binding protein (Figure 3C), or alternatively RBP can be added directly in the buffer channel (Figure 3B). - To capture the beads, move the optical traps apart from each other. First move to the AD channel and catch an AD-bead in trap 1. Next, move the stage to the SA-channel and catch a single SA bead by trap 2.
NOTE: Try to stay at the interface of the buffer and bead channels to avoid losing the already caught bead, or to prevent catching multiple beads by the same trap. - Once the beads of the right size are captured, move to the buffer channel and stop the laminar flow. Next, perform force calibration to check trap stiffness. The respective stiffness values should not differ in the x/y axis by more than 10-15%.
NOTE: Adjust the laser power or the laser split between the traps according to bead size. Force calibration does not have to be done for every bead pair as long as the bead templates match (similarity score > 0.9). However, it should be performed regularly, or at least every time assay conditions are changed. - Start fishing for a tether by moving the beads close to each other, waiting for a few seconds, and then moving them back apart, repeat until a tether is formed. A tether formation results in an increase of measured force upon pulling the two beads away from each other.
NOTE: To avoid formation of multiple tethers, the beads should not be moved too close. Upon catching a tether between the two beads, tether quality can be checked by finding the overstretching plateau. The plateau should be between 50 to 60 pN for a single tether. - Upon obtaining a tether, start the measurement. Depending on the phenomenon studied different measurement setups should be chosen (Figure 1B-D).
NOTE: Usually at the beginning of the experiment, a force-ramp experiment is conducted to check the tether quality and probe the behavior. Afterward, one may also start the constant-force or constant-position experiments to study the state transitions further. Once sufficient number of measurements have been performed on an RNA sample to determine its behavior, labeled factors can be added to the system to perform confocal measurements. - To perform fluorescence measurements, turn on the confocal lasers and photon counter unit in the optical tweezers instrument.
- Turn on the excitation laser of desired wavelength in the software interface and set the power of the laser to 5% or higher, depending on the fluorophore.
NOTE: While not measuring lower the power setting of the excitation laser to 0% to avoid excessive photodamage to the sample. - Start imaging the sample by using image functions of the software.
NOTE: In order to get well-focused images, the focal plane of the confocal microscope and optical traps have to be aligned. For this purpose, autofluorescence of the polystyrene beads in the blue laser channel can be employed. The focal plane of optical traps is moved up or down in the z-axis until the image of beads reaches its highest diameter. At this position, the fluorescence signal from the molecule tethered between the beads can be measured. - To use the kymograph function, specify the x-y position of the kymograph axis so that it allows detection of the tether between the beads.
- Throughout the measurement, buffer composition can be easily changed by either moving the beads to different channels or by changing the buffer supplied in the microfluidics system.
4. Data analysis
- Raw data pre-processing
- By using a simple script, downsample the raw data enough to (i) allow faster subsequent data processing but (ii) still contain all the critical information (Figure 4A). Usually, 100-5000 Hz is suitable for this purpose.
NOTE: The data gathering frequency in optical tweezers experiments is often higher than it is necessary for the analysis - in the presented experiments, the data gathering frequency is set to 78 125 Hz by default. Since storage space is limited, it is convenient and timesaving to reduce the sampling rate of the data. Here, the raw data were downsampled by a factor of 30. - Next, employ a signal filter to reduce the high frequency measurement noise from the signal (Figure 4A). Adjust the filter degree and cut-off frequency parameters accordingly to optimize data output of different experiments (Figure 5).
NOTE: Amongst signal filters, Butterworth filter51 is one of the most widely used. A custom-written python script allowing the pre-processing of raw data is provided in the supplementary data. Downsampling and signal filtering parameters (cut-off frequency, filter degree) need to be optimized for different experiments.
- By using a simple script, downsample the raw data enough to (i) allow faster subsequent data processing but (ii) still contain all the critical information (Figure 4A). Usually, 100-5000 Hz is suitable for this purpose.
- For force-ramp data analysis, use the following steps.
- Mark the steps either manually by finding corresponding points on the force trajectory plot or by using custom-written scripts. Unfolding steps are characterized by a sudden drop in force combined with an increase in distance in the force-distance (FD) curve.
- Once unfolding events are marked, fit different regions of the FD curve using appropriate models (Figure 4D).
NOTE: For the region before the first unfolding step, the tether can be considered "double-stranded" and is commonly fit using an extensible Worm-like-chain model (WLC)47,52,53. The parts after the first unfolding event are considered a combination of double-stranded nucleotides (handles) and single-stranded nucleotides (unfolded RNA molecule). Therefore, data fitting is more complex - usually a combination of 2 WLC models or WLC and Freely-jointed chain (FJC) models are employed36,39,52. The extensible WLC model has two main fit parameters the contour length (LC) and the persistence length (LP). Contour length corresponds to the length of the fully stretched molecule and persistence length defines the bending properties of the molecule of interest. The model can be described with the following equation (1). WLC can be used to model the behavior of both folded as well as unfolded regions, although for each of these a separate model with different parameters has to be employed.
(1)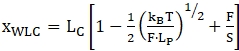
where x is extension, LC is contour length, F is force, LP is persistence length, kB is Boltzmann constant, T is Thermodynamic temperature, and S is stretch modulus.
The second model called Freely-jointed chain (FJC) is commonly used to describe behavior of unfolded single stranded regions. It uses similar parameters of the polymers but treats each unit of the "chain" as a rigid rod, here corresponding to the nucleotides of the unfolded single stranded region. The following equation (2) describes this model:
(2)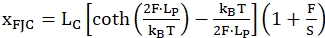
NOTE: Our lab has recently developed an algorithm that allows batch processing of the raw force-ramp data called "Practical Optical Tweezers Analysis TOol (POTATO)54." The algorithm downsamples and filters the data, then it identifies possible unfolding steps and finally performs data fitting. The POTATO is built in a user-friendly graphical user interface (GUI) (https://github.com/REMI-HIRI/POTATO).
- Process constant-force data as follows:
NOTE: The following instructions can be analogically applied on constant-position data.- For the constant-force data, plot the distance over time (Figure 5). A histogram showing the frequency (counts) of different conformations over the relative change in position is a useful way to characterize various dominant and minor states (Figure 7).
- Fit the histogram using (multiple) Gaussian functions to estimate the overall percentage of individual conformers at a given force (Figure 7C). The Gaussian fits, mean position, and the standard deviation outlines the force-related relationship among different populations.
NOTE: A custom-written python script allowing pre-processing and basic bimodal Gaussian fitting of constant-force data is provided in the supplementary data. Parameters (cut-off frequency, filter degree, expected means, standard deviation values and amplitudes) need to be optimized for different experiments. - Next, employ the Hidden Markov model to further analyze the states, which may uncover additional folding intermediates (conformers)55. For further information on the constant-force and Hidden Markov model, one may refer to55,56,57,58.
Results
In this section, focus is mainly given on measurements of RNA-protein/ligand interactions by the fluorescence optical tweezers. For a description of general RNA optical tweezers experiments and corresponding representative results, see32. For more detailed discussion of the RNA/DNA-protein interactions, also see1,2,26,59,60.
Discussion
Here, we demonstrate the use of fluorescence-coupled optical tweezers to study interactions and dynamic behavior of RNA molecules with various ligands. Below, critical steps and limitations of the present technique are discussed.
Critical steps in the protocol
As for many other methods, the quality of the sample is pivotal to obtain reliable data. Therefore, to obtain the highest possible quality samples, it is worth it to spend time to optimize the procedure for sample ...
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Anuja Kibe and Jun. Prof. Redmond Smyth for critically reviewing the manuscript. We thank Tatyana Koch for expert technical assistance. We thank Kristyna Pekarkova for the help with recording experimental videos. The work in our laboratory is supported by the Helmholtz Association and funding from the European Research Council (ERC) Grant Nr. 948636 (to NC).
Materials
| Name | Company | Catalog Number | Comments |
| Bacterial Strains | |||
| E. coli HB101 | lab collection | N/A | cloning of the vectors |
| Chemicals and enzymes | |||
| Sodium chloride | Sigma-Aldrich | 31424 | Buffers |
| Biotin-16-dUTP | Roche | 11093070910 | Biotinylation |
| BSA | Sigma-Aldrich | A4737 | Buffers |
| Catalase | Lumicks | N/A | Oxygen scavanger system |
| Dithiothreitol (DTT) | Melford Labs | D11000 | Buffers |
| DNAse I from bovine pancreas | Sigma-Aldrich | D4527 | in vitro transcription |
| dNTPs | Th.Geyer | 11786181 | PCR |
| EDTA | Sigma-Aldrich | E9884 | Buffers |
| Formamide | Sigma-Aldrich | 11814320001 | Buffers |
| Glucose | Sigma-Aldrich | G8270-1KG | Oxygen scavanger system |
| Glucose-oxidase | Lumicks | N/A | Oxygen scavanger system |
| HEPES | Carl Roth | HN78.3 | Buffers |
| Magnesium chloride | Carl Roth | 2189.1 | Buffers |
| Phusion DNA polymerase | NEB | M0530L | Gibson assembly, cloning |
| Potassium chloride | Merck | 529552-1KG | Buffers |
| PrimeSTAR GXL DNA Polymerase | Takara Bio Clontech | R050A | PCR |
| Pyrophosphotase, thermostabile, inorganic | NEB | M0296L | in vitro transcription |
| RNase Inhibitor | Molox | 1000379515 | Buffers |
| rNTPS | life technologies | R0481 | in vitro transcription |
| Sodium thiosulophate | Sigma-Aldrich | S6672-500G | Bleach deactivation |
| Sytox Green | Lumicks | N/A | confocal measurements |
| T4 DNA Polymerase | NEB | M0203S | Biotinylation |
| T5 exonuclease | NEB | M0363S | Gibson assembly, cloning |
| T7 RNA polymerase | Produced in-house | N/A | in vitro transcription |
| Taq DNA polymerase | NEB | M0267S | PCR |
| Taq ligase | Biozym | L6060L | Gibson assembly, cloning |
| TWEEN 20 BioXtra | Sigma-Aldrich | P7949 | Buffers |
| Kits | |||
| Monolith Protein Labeling Kit RED-NHS 2nd Generation (Amine Reactive) | Nanotemper | MO-L011 | Used for ribosome labeling |
| Purefrex 2.0 | GeneFrontier | PF201-0.25-EX | Ribosomes used for the labeling |
| Oligonucleotides | |||
| 5' handle T7 forward | Microsynth | custom order | 5’ - CTTAATACGACTCACTATAGGTC CTTTCTGTGGACGCC - 3’, used to generate OT in vitro transcription template in PCR 1 |
| 3’ handle reverse | Microsynth | custom order | 5' - GTCAAAGTGCGCCCCGTTATCC - 3', used to generate OT in vitro transcription template in PCR 1 |
| 5' handle forward | Microsynth | custom order | 5' - TCCTTTCTGTGGACGCCGC - 3' , used to generate 5' handle in PCR 2 |
| 5’ handle reverse | Microsynth | custom order | 5’ - CATAAATACCTCTTTACTAATATA TATACCTTCGTAAGCTAGCGT - 3’, used to generate 5' handle in PCR 2 |
| 3’ handle forward | Microsynth | custom order | 5' - ATCCTGCAACCTGCTCTTCGCC AG - 3', used to generate 3' handle in PCR 3 |
| 3’ handle reverse 5’labeled with digoxigenin | Microsynth | custom order | 5' -[Dig]-GTCAAAGTGCGCCCCGTTATCC - 3', used to generate 3' handle in PCR 3 |
| DNA vectors | |||
| pMZ_OT | produced in-house | N/A | further description in "Structural studies of Cardiovirus 2A protein reveal the molecular basis for RNA recognition and translational control" Chris H. Hill, Sawsan Napthine, Lukas Pekarek, Anuja Kibe, Andrew E. Firth, Stephen C. Graham, Neva Caliskan, Ian Brierley bioRxiv 2020.08.11.245035; doi: https://doi.org/10.1101/2020.08.11.245035 |
| Software and Algorithms | |||
| Atom | https://atom.io/packages/ide-python | N/A | |
| Bluelake | Lumicks | N/A | |
| Graphpad | https://www.graphpad.com/ | N/A | |
| InkScape 0.92.3 | https://inkscape.org/ | N/A | |
| Matlab | https://www.mathworks.com/products/matlab.html | N/A | |
| POTATO | https://github.com/lpekarek/POTATO.git | N/A | |
| RNAstructure | https://rna.urmc.rochester.edu/RNAstructure.html | N/A | |
| Spyder | https://www.spyder-ide.org/ | N/A | |
| Other | |||
| Streptavidin Coated Polystyrene Particles, 1.5-1.9 µm, 5 ml, 1.0% w/v | Spherotech | SVP-15-5 | |
| Anti-digoxigenin Coated Polystyrene Particles, 2.0-2.4 µm, 2 ml, 0.1% w/v | Spherotech | DIGP-20-2 | |
| Syringes | VWR | TERUMO SS+03L1 | |
| Devices | |||
| C-trap | Lumicks | N/A | optical tweezers coupled with confocal microscopy |
References
- Balcerak, A., Trebinska-Stryjewska, A., Konopinski, R., Wakula, M., Grzybowska, E. A. RNA-protein interactions: disorder, moonlighting and junk contribute to eukaryotic complexity. Open Biology. 9 (6), 190096 (2019).
- Armaos, A., Zacco, E., Sanchez de Groot, N., Tartaglia, G. G. RNA-protein interactions: Central players in coordination of regulatory networks. BioEssays. 43 (2), 2000118 (2021).
- Firth, A. E., Brierley, I. Non-canonical translation in RNA viruses. Journal of General Virology. 93, 1385-1409 (2012).
- Caliskan, N., Peske, F., Rodnina, M. V. Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends in Biochemical Sciences. 40 (5), 265-274 (2015).
- Jaafar, Z. A., Kieft, J. S. Viral RNA structure-based strategies to manipulate translation. Nature Reviews Microbiology. 17 (2), 110-123 (2019).
- Eswarappa, S. M., et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 157 (7), 1605-1618 (2014).
- Rodnina, M. V., et al. Translational recoding: canonical translation mechanisms reinterpreted. Nucleic Acids Research. 48 (3), 1056-1067 (2020).
- Li, Y., et al. Transactivation of programmed ribosomal frameshifting by a viral protein. Proceedings of the National Academy of Sciences. 111 (21), 2172 (2014).
- Napthine, S., et al. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nature Communications. 8 (1), 15582 (2017).
- Patel, A., et al. Molecular characterization of the RNA-protein complex directing -2/-1 programmed ribosomal frameshifting during arterivirus replicase expression. Journal of Biological Chemistry. 295 (52), 17904-17921 (2020).
- Napthine, S., Bell, S., Hill, C. H., Brierley, I., Firth, A. E. Characterization of the stimulators of protein-directed ribosomal frameshifting in Theiler's murine encephalomyelitis virus. Nucleic Acids Research. 47 (15), 8207-8223 (2019).
- Marshall, R. A., Aitken, C. E., Dorywalska, M., Puglisi, J. D. Translation at the Single-Molecule Level. Annual Review of Biochemistry. 77 (1), 177-203 (2008).
- Rodnina, M. V. The ribosome in action: Tuning of translational efficiency and protein folding. Protein science : A publication of the Protein Society. 25 (8), 1390-1406 (2016).
- Rodnina, M. V., Fischer, N., Maracci, C., Stark, H. Ribosome dynamics during decoding. Philosophical Transactions of Royal Society of London B Biological Sciences. 372 (1716), (2017).
- Yan, S., Wen, J. D., Bustamante, C., Tinoco, I. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 160 (5), 870-881 (2015).
- Liu, T., et al. Direct measurement of the mechanical work during translocation by the ribosome. eLife. 3, 03406 (2014).
- Desai, V. P., et al. Co-temporal force and fluorescence measurements reveal a ribosomal gear shift mechanism of translation regulation by structured mRNAs. Molecular Cell. 75 (5), 1007-1019 (2019).
- Choi, J., O'Loughlin, S., Atkins, J. F., Puglisi, J. D. The energy landscape of -1 ribosomal frameshifting. Science Advances. 6 (1), (2020).
- Prabhakar, A., Puglisi, E. V., Puglisi, J. D. Single-molecule fluorescence applied to translation. Cold Spring Harbor Perspectives in Biology. 11 (1), 032714 (2019).
- Bao, C., et al. mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding. Elife. 9, 55799 (2020).
- Chen, J., Tsai, A., O'Leary, S. E., Petrov, A., Puglisi, J. D. Unraveling the dynamics of ribosome translocation. Current Opinion in Structural Biology. 22 (6), 804-814 (2012).
- Qu, X., et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 475 (7354), 118-121 (2011).
- Zheng, Q., et al. Ultra-stable organic fluorophores for single-molecule research. Chemical Society Reviews. 43 (4), 1044-1056 (2014).
- Blanchard, S. C. Single-molecule observations of ribosome function. Current Opinion in Structural Biology. 19 (1), 103-109 (2009).
- Juette, M. F., et al. The bright future of single-molecule fluorescence imaging. Current Opinion in Chemical Biology. 20, 103-111 (2014).
- McCauley, M. J., Williams, M. C. Mechanisms of DNA binding determined in optical tweezers experiments. Biopolymers. 85 (2), 154-168 (2007).
- Ashkin, A., Dziedzic, J. M., Bjorkholm, J. E., Chu, S. Observation of a single-beam gradient force optical trap for dielectric particles. Optics Letters. 11 (5), 288-290 (1986).
- Bustamante, C., Smith, S. B., Liphardt, J., Smith, D. Single-molecule studies of DNA mechanics. Current Opinion in Structural Biology. 10 (3), 279-285 (2000).
- Choudhary, D., Mossa, A., Jadhav, M., Cecconi, C. Bio-molecular applications of recent developments in optical tweezers. Biomolecules. 9 (1), 23 (2019).
- Moffitt, J. R., Chemla, Y. R., Smith, S. B., Bustamante, C. Recent advances in optical tweezers. Annual Reviews of Biochemistry. 77, 205-228 (2008).
- Li, P. T. X., Vieregg, J., Tinoco, I. How RNA Unfolds and Refolds. Annual Review of Biochemistry. 77 (1), 77-100 (2008).
- Stephenson, W., Wan, G., Tenenbaum, S. A., Li, P. T. Nanomanipulation of single RNA molecules by optical tweezers. Journal of Visualized Experiments. (90), e51542 (2014).
- Halma, M. T. J., Ritchie, D. B., Cappellano, T. R., Neupane, K., Woodside, M. T. Complex dynamics under tension in a high-efficiency frameshift stimulatory structure. Proceedings of the National Academy of Sciences. 116 (39), 19500 (2019).
- Hansen, T. M., Reihani, S. N. S., Oddershede, L. B., Sørensen, M. A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proceedings of the National Academy of Sciences of the United States of America. 104 (14), 5830-5835 (2007).
- Zhong, Z., et al. Mechanical unfolding kinetics of the SRV-1 gag-pro mRNA pseudoknot: possible implications for -1 ribosomal frameshifting stimulation. Science Reports. 6, 39549 (2016).
- McCauley, M. J., Rouzina, I., Li, J., Núñez, M. E., Williams, M. C. Significant differences in RNA structure destabilization by HIV-1 GagDp6 and NCp7 proteins. Viruses. 12 (5), 484 (2020).
- de Messieres, M., et al. Single-molecule measurements of the CCR5 mRNA unfolding pathways. Biophysics Journal. 106 (1), 244-252 (2014).
- Yang, L., et al. Single-molecule mechanical folding and unfolding of RNA hairpins: Effects of single A-U to A·C pair substitutions and single proton binding and implications for mRNA structure-induced -1 ribosomal frameshifting. Journal of American Chemical Society. 140 (26), 8172-8184 (2018).
- McCauley, M. J., et al. Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA. Proceedings of the National Academy of Sciences of the United States of America. 112 (44), 13555-13560 (2015).
- Whitley, K. D., Comstock, M. J., Chemla, Y. R. High-resolution "Fleezers": Dual-trap optical tweezers combined with single-molecule fluorescence detection. Methods in Molecular Biology. 1486, 183-256 (2017).
- Yerramilli, V. S., Kim, K. H. Labeling RNAs in live cells using malachite green aptamer scaffolds as fluorescent probes. ACS Synthetic Biology. 7 (3), 758-766 (2018).
- Gross, P., Farge, G., Peterman, E. J., Wuite, G. J. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods in Enzymology. 475, 427-453 (2010).
- Zimmer, M. M., et al. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nature Communications. 12 (1), 7193 (2021).
- Neupane, K., Yu, H., Foster, D. A. N., Wang, F., Woodside, M. T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic acids research. 39 (17), 7677-7687 (2011).
- Ritchie, D. B., Soong, J., Sikkema, W. K., Woodside, M. T. Anti-frameshifting ligand reduces the conformational plasticity of the SARS virus pseudoknot. Journal of the American Chemical Society. 136 (6), 2196-2199 (2014).
- Janissen, R., et al. Invincible DNA tethers: covalent DNA anchoring for enhanced temporal and force stability in magnetic tweezers experiments. Nucleic Acids Research. 42 (18), 137 (2014).
- Smith, S. B., Cui, Y., Bustamante, C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271 (5250), 795 (1996).
- Puljung, M. C., Zagotta, W. N. Labeling of specific cysteines in proteins using reversible metal protection. Biophysical Journal. 100 (10), 2513-2521 (2011).
- Toseland, C. P. Fluorescent labeling and modification of proteins. Journal of Chemical Biology. 6 (3), 85-95 (2013).
- Hill, C. H., et al. Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch. Nature Communications. 12 (1), 7166 (2021).
- Butterworth, S. On the theory of filter amplifiers. Experimental Wireless and the Wireless Engineer. 7, 536-541 (1930).
- Wang, M. D., Yin, H., Landick, R., Gelles, J., Block, S. M. Stretching DNA with optical tweezers. Biophysics Journal. 72 (3), 1335-1346 (1997).
- Mukhortava, A., et al. Structural heterogeneity of attC integron recombination sites revealed by optical tweezers. Nucleic Acids Research. 47 (4), 1861-1870 (2019).
- Buck, S., Pekarek, L., Caliskan, N. POTATO: An automated pipeline for batch analysis of optical tweezers data. bioRxiv. , (2021).
- Zhang, Y., Jiao, J., Rebane, A. A. Hidden Markov modeling with detailed balance and its application to single protein folding. Biophysical Journal. 111 (10), 2110-2124 (2016).
- Sgouralis, I., Pressé, S. An introduction to infinite HMMs for single-molecule data analysis. Biophysics Journal. 112 (10), 2021-2029 (2017).
- Müllner, F. E., Syed, S., Selvin, P. R., Sigworth, F. J. Improved hidden Markov models for molecular motors, part 1: basic theory. Biophysical Journal. 99 (11), 3684-3695 (2010).
- Elms, P. J., Chodera, J. D., Bustamante, C. J., Marqusee, S. Limitations of constant-force-feedback experiments. Biophysical Journal. 103 (7), 1490-1499 (2012).
- Re, A., Joshi, T., Kulberkyte, E., Morris, Q., Workman, C. T. RNA-protein interactions: an overview. Methods Molecular Biology. 1097, 491-521 (2014).
- Jankowsky, E., Harris, M. E. Specificity and nonspecificity in RNA-protein interactions. Nature reviews. Molecular Cell Biology. 16 (9), 533-544 (2015).
- Lim, F., Peabody, D. S. RNA recognition site of PP7 coat protein. Nucleic Acids Research. 30 (19), 4138-4144 (2002).
- Sunbul, M., Jäschke, A. SRB-2: a promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Research. 46 (18), 110 (2018).
- Sanchez de Groot, N., et al. RNA structure drives interaction with proteins. Nature Communications. 10 (1), 3246 (2019).
- Zeffman, A., Hassard, S., Varani, G., Lever, A. The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the Gag polyprotein. Journal of Molecular Biology. 297 (4), 877-893 (2000).
- Mangeol, P., et al. Probing ribosomal protein-RNA interactions with an external force. Proceedings of the National Academy of Sciences. 108 (45), 18272 (2011).
- Luo, X., et al. Molecular mechanism of RNA recognition by Zinc-Finger antiviral protein. Cell Reports. 30 (1), 46-52 (2020).
- Qu, X., Lancaster, L., Noller, H. F., Bustamante, C., Tinoco, I. Ribosomal protein S1 unwinds double-stranded RNA in multiple steps. Proceedings of the National Academy of Science U. S. A. 109 (36), 14458-14463 (2012).
- Chandra, V., Hannan, Z., Xu, H., Mandal, M. Single-molecule analysis reveals multi-state folding of a guanine riboswitch. Nature Chemical Biology. 13 (2), 194-201 (2017).
- Savinov, A., Perez, C. F., Block, S. M. Single-molecule studies of riboswitch folding. Biochimica et Biophysica Acta. 1839 (10), 1030-1045 (2014).
- Kelly, J. A., et al. Structural and functional conservation of the programmed ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). Journal of Biological Chemistry. 295 (31), 10741-10748 (2020).
- Neupane, K., et al. Structural dynamics of single SARS-CoV-2 pseudoknot molecules reveal topologically distinct conformers. Nature Communications. 12 (1), 4749 (2021).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Zheng, Q., Jockusch, S., Zhou, Z., Blanchard, S. C. The contribution of reactive oxygen species to the photobleaching of organic fluorophores. Photochemistry and Photobiology. 90 (2), 448-454 (2014).
- Deerinck, T. J. The application of fluorescent quantum dots to confocal, multiphoton, and electron microscopic imaging. Toxicologic Pathology. 36 (1), 112-116 (2008).
- Rill, N., Mukhortava, A., Lorenz, S., Tessmer, I. Alkyltransferase-like protein clusters scan DNA rapidly over long distances and recruit NER to alkyl-DNA lesions. Proceedings of the National Academy of Science U. S. A. 117 (17), 9318-9328 (2020).
- Swoboda, M., et al. Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments. ACS Nano. 6 (7), 6364-6369 (2012).
- Aitken, C. E., Marshall, R. A., Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophysical Journal. 94 (5), 1826-1835 (2008).
- Wen, J. -. D., et al. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophysical journal. 92 (9), 2996-3009 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
ISSN 2578-2037
Copyright © 2025 MyJoVE Corporation. All rights reserved
We use cookies to enhance your experience on our website.
By continuing to use our website or clicking “Continue”, you are agreeing to accept our cookies.