Method Article
Culturing and Screening the Plant Parasitic Nematode Ditylenchus dipsaci
In This Article
Summary
The present protocol describes a reliable and straightforward method for culturing, collecting, and screening Ditylenchus dipsaci.
Abstract
Plant-parasitic nematodes (PPNs) destroy over 12% of global food crops every year, which equates to roughly 157 billion dollars (USD) lost annually. With a growing global population and limited arable land, controlling PPN infestation is critical for food production. Compounding the challenge of maximizing crop yields are the mounting restrictions on effective pesticides because of a lack of nematode selectivity. Hence, developing new and safe chemical nematicides is vital to food security. In this protocol, the culture and collection of the PPN species Ditylenchus dipsaci are demonstrated. D. dipsaci is both economically damaging and relatively resistant to most modern nematicides. The current work also explains how to use these nematodes in screens for novel small molecule nematicides and reports on data collection and analysis methodologies. The demonstrated pipeline affords a throughput of thousands of compounds per week and can be easily adapted for use with other PPN species such as Pratylenchus penetrans. The techniques described herein can be used to discover new nematicides, which may, in turn, be further developed into highly selective commercial products that safely combat PPNs to help feed an increasingly hungry world.
Introduction
Plant-parasitic nematodes (PPNs) are estimated to be responsible for the loss of 12.3% of global food production and cause an estimated 157 billion dollars in damage annually1,2,3. Unfortunately, the ability to control PPNs is waning because effective chemical nematicides have been banned or are facing escalating restrictions because of human safety and environmental concerns. This is primarily due to the poor nematode selectivity of previous generations of pesticides4. Over the last 25 years, six new chemical nematicides have been piloted or introduced into the market5. One of these has already been banned in Europe, and another has been discontinued while being investigated for its impact on human heatlh6,7. Hence, there is a pressing need for new nematicides that are highly selective for PPNs.
The stem and bulb nematode, Ditylenchus dipsaci (D. dipsaci) is an economically impactful PPN4. D. dipsaci infects nearly 500 plant species across 30 biological races and targets some of the most agriculturally important crops such rye, oats, garlic, onion, and leek8,9. For example, D. dipsaci has recently scourged garlic fields in Ontario and Quebec, resulting in losses of up to 90%10,11. Its geographical distribution is nearly ubiquitous and includes the Americas (including California and Florida), Europe, much of Asia (including China), and Oceania9. D. dipsaci is a migratory endoparasite that enters the stomata on leaves or wounds and lenticels where they release enzymes to break down the cell wall12. Compounding the impact of D. dipsaci on crops, the damage caused by the PPN makes the plant susceptible to secondary infection11. Unfortunately, D. dipsaci shows high tolerance levels to current nematicides compared to other nematode strains13,14.
This protocol describes the culturing of D. dipsaci, and its use in large-scale screens for small molecule candidate nematicides. Briefly, D. dipsaci populations are maintained and expanded on pea plants cultured in sterile Gamborg B-5 (GA) media15. Before growing seed sprouts on GA medium, the seeds must be sterilized through a series of washes and plated on nutrient agar (NA) to check for contamination. Seed sterilization is essential to detect bacterial and fungal contaminants that may be present. The non-contaminated seeds are then transferred to GA plates, where seed sprouts will grow in preparation for infection. The GA plates containing seed sprouts are infected with nematodes from a previous culture plate by transferring a piece of agar containing root tissue to the fresh plates. After 6-8 weeks, the nematodes are extracted from the GA media and are filtered through a coffee filter-lined funnel into a collection beaker. The nematodes can be used in various bioassays once a suitable number has been collected. The technique described in this protocol generates approximately 15,000 D. dipsaci per culture plate. Alternative protocols to cultivate D. dipsaci have been published16,17.
An in vitro small molecule screening assay based on previous work18 is also described here. As a proxy of worm health, the mobility of 20 nematodes per well is examined after 5 days of small molecule exposure. To better visualize worm mobility, NaOH is added to increase the movement of live worms19,20. This protocol allows medium-throughput screening and provides valuable data to assess the nematicidal potential of small molecules. If a different nematode collection technique is used16,17, the small molecule screening methodology described herein can nevertheless be implemented.
Protocol
The D. dipsaci strain G-137 used for the present work was collected from Fish Lake 4 Variety garlic in Prince Edward County and was provided by Agriculture and Agri-food Canada. If starting a fresh culture, consult Poirier et al. for inoculation methodology15.
1. Culturing of D. dipsaci
- Prepare the media and plates following previously reported work15.
- Prepare 500 mL of Nutrient agar (NA) media with 23 g/L of NA (see Table of Materials) and ultrapure water. Using sterile technique, pour 25 mL of autoclaved NA media into 20 disposable Petri dishes (100 mm diameter x 15 mm deep). Allow agar to solidify at room temperature (22 °C) with the lid on for ~2 h and set aside for later use.
- Prepare 500 mL of Gamborg B-5 (GA) media containing 3.2 g/L of GA basal medium with minimal organics, 20 g/L of sucrose, 15 g/L of agar, and distilled water (see Table of Materials). Using sterile technique, pour 50 mL of autoclaved GA media in 10 disposable Petri dishes (100 mm diameter x 25 mm deep). Allow agar to solidify at room temperature (22 °C) with the lid on for ~5 h and store upright at room temperature in a sterile bag until the sprout transfer.
NOTE: Excess media plates are made in case contamination occurs.
- Perform seed sterilization following a method modified from a previous work15.
- Autoclave one 2 L beaker with a stir bar, 2 forceps, a glass petri dish, and 1 L of distilled water. Prepare 200 mL of 95% EtOH solution and 200 mL of a 15% commercial bleach solution.
- Use pea seeds (see Table of Materials). Pour 150 seeds into a sterile 2 L beaker with a stir bar near a Bunsen burner flame on a lab bench.
- Add 200 mL of 95% EtOH to the seeds within the beaker, stir vigorously on the stir plate for 5 min, and then pour off EtOH in a waste container.
- Pour bleach solution into the beaker to completely immerse the seeds. Stir vigorously on a stir plate for 20 min then pour off bleach in a waste container.
- Pour distilled water into the beaker to immerse seeds and stir vigorously on a stir plate for 20 min. Repeat water washes three times, pouring off distilled water after each wash. After the final water wash, pour sterilized seeds into the glass Petri dish.
- To check for contamination, transfer 6 seeds to each 10 cm NA plate (prepared in step 1.1.1) in the laminar flood hood using sterilized forceps. Arrange the seeds around the plate's circumference (plumper seeds work best). Wrap the plates individually in laboratory wrapping film and incubate in the dark for 3 days at 26 °C.
NOTE: Plating more seeds than needed will allow for selective use of non-contaminated seeds.
- Perform sprout transfer following the step below.
- In a laminar flow hood, use sterilized forceps to plate 2 non-contaminated seeds on each GA plate (prepared in step 1.1.2). Wrap the plates individually in laboratory wrapping film and incubate at room temperature for 7-10 days to allow seeds to sprout.
- Perform in vitro rearing following a method modified from a previous work15.
- Prepare 50 mL of 20 g/L sucrose solution. Filter sterilize sucrose solution and set aside.
- In a laminar flow hood, cut a piece of agar containing root tissue (~2 cm3) from an existing culture plate. Pipette 500 µL of sucrose solution on new GA plate with pea seedlings and place agar cube on top of sucrose. Wrap the plates individually in laboratory wrapping film and maintain the culture in a box lined with aluminum foil at room temperature (~22 °C).
- Subculture nematodes on fresh GA plates every 8-9 weeks to maintain the culture. Nematodes are ready to be extracted after ~8 weeks.
2. Extraction and collection of D. dipsaci
- Perform the extraction of D. dipsaci following the steps below.
- Autoclave a 50 mL beaker, an 80 mm funnel, 150 mL distilled water, and coffee filters. Place the funnel into the beaker and line the funnel with sterile coffee filter.
- In a laminar flow hood, cut the agar and the root tissue into 1 cm3 using a sterile scalpel. Transfer the agar cubes into the coffee filter-lined funnel and slowly pour distilled water on the agar to moisten the coffee filter.
- Remove the coffee filter-lined funnel from the beaker and fill the beaker with distilled water until the water level is just touching the bottom of the filter once the coffee filter-lined funnel is replaced.
- Cover the coffee filter-lined funnel and beaker with aluminum foil. Leave overnight (16 h) on the benchtop to allow worms to move through the coffee filter into the collection beaker.
NOTE: A nematode-culture plate is ready for extraction when worms have crawled into the agar when examined with a dissection microscope. Typically, a plate is ready for extraction 6-8 weeks after its initial inoculation.
- Perform the collection of D. dipsaci.
NOTE: The next day, the D. dipsaci worms will have settled to the bottom of the beaker.- Remove the coffee filter-lined funnel and aspirate the top 40 mL of water from the collection beaker; ensure not to disrupt the settled worms. Using a 10 mL plastic serological pipette, collect the remaining liquid into a 15 mL conical centrifuge tube.
NOTE: This collection can be used directly in assays. Place at 4 °C if they will not be used within 24 hours. The worms can be left for 3 days at 4 °C with no visible impact on mobility. Removing the funnel may disturb the worms in the collection beaker. Let them settle to the bottom before collecting.
- Remove the coffee filter-lined funnel and aspirate the top 40 mL of water from the collection beaker; ensure not to disrupt the settled worms. Using a 10 mL plastic serological pipette, collect the remaining liquid into a 15 mL conical centrifuge tube.
3. In vitro small molecule screen
- Prepare the assay plates following the steps below.
- Pour autoclaved distilled water into a sterile trough and, using a multichannel pipette, dispense 40 µL of distilled water from the trough into each well of a flat-bottom 96-well plate.
- Prepare pinning tool and add the chemicals
- Set up pinner trays (see Table of Materials) near a Bunsen burner flame on a lab bench. Add the following to successive pinner trays: 25 mL of pin cleaning solution, 35 mL of 50% DMSO (in water), 45 mL of distilled water, 55 mL of 70% EtOH, and 65 mL of 95% EtOH. Place one piece of blotting paper in front of each tray
- Clean the pinning tool by emersing the pins in the cleaning solution and moving the pins up and down three times (3x) in the solution. In this protocol, '3x' and '10x' are defined as moving the pinner up and down in a solution three or ten times, respectively. Blot the pins on the blotting paper. Repeat this procedure once more.
- Next, rinse the pins 3x in distilled water, followed by blotting the pins. Repeat the procedure once more.
- Lastly, rinse the pins 3x in 95% EtOH, followed by blotting the pins. Repeat once more. Flame the pinner and allow the ethanol to evaporate.
- Add chemicals from the 96-well chemical stock plates to assay plates by pinning 3x into the chemical plate, then transferring the pins 10X into the assay plate. Blot onto paper in front of the cleaning solution.
- Clean pinning tool between plates by washing in the following order, blotting in between on blotting paper: 3x in 50% DMSO (once), 3x in distilled water (once), 3x in 75% EtOH (once), 3x in 95% EtOH (twice). Flame the pinner and allow ethanol to evaporate.
- Repeat step 3.2.2 when all pinning is completed.
NOTE: The screening was performed at a final concentration of 60 µM.
- Addition of worms
- Count the number of nematodes from the collection by first resuspending and then pipetting 5 µL using low retention tips onto a slide for observation. Count the number of nematodes in 5 µL using a dissection microscope.
- Adjust the concentration to 2 worms/µL using sterile distilled water. Using a multichannel pipette and a trough, add 10 µL (~20 worms) to each well of the 96-well plates.
NOTE: Approximately 15,000 D. dipsaci nematodes will be collected per culture plate using the described culture and collection method. Twenty worms are used per well because the small number facilitates the clear visualization and accounting of mobile and immobile ones.
- Adjust the concentration to 2 worms/µL using sterile distilled water. Using a multichannel pipette and a trough, add 10 µL (~20 worms) to each well of the 96-well plates.
- Seal the plates in laboratory wrapping film and wrap with a damp paper towel. Place in box and affix on a sticky pad in 20 °C shaking incubator set at 200 rpm. Ensure that plates are stabilized in the box by adding an extra damp paper towel to ensure minimal movement of plates.
- Count the number of nematodes from the collection by first resuspending and then pipetting 5 µL using low retention tips onto a slide for observation. Count the number of nematodes in 5 µL using a dissection microscope.
4. Data collection and analysis
- Observe plates on day 5 under a dissecting microscope. Count the number of mobile and total number of D. dipsaci in DMSO solvent controls and drug-treated wells.
- If the worms are relatively immobile, add 2 µL of 1 M NaOH to a final concentration of 40 mM to the well to stimulate movement19, 20.
NOTE: After adding NaOH, worms will move instantly and need to be viewed within 5 min. The number of mobile worms and the total number of worms will be used to calculate the proportion of mobile worms. The length of the assay may change depending on the aim of the screen.
- If the worms are relatively immobile, add 2 µL of 1 M NaOH to a final concentration of 40 mM to the well to stimulate movement19, 20.
- Calculate the proportion of mobile worms. In the D. dipsaci screens, wells that reproducibly yielded 0% mobile worms are categorized as strong hits.
NOTE: Prolonged exposure of plates on the stage of dissection microscopes is avoided because the light source can heat the plates and induce variable effects on the bioassay.
Results
Independent replicates reveal reproducible hits
To illustrate the expected variation between the replicate screens, the means and variation in sample mobility are plotted from three representative plates from a recent screen (Figure 1). Three replicates of the screen were performed on three different days. All three plates have negative (solvent-only) controls (darker bars), and samples within the set contained established nematicides. The remaining compounds are from a custom library currently being characterized in the Roy lab. Compared to similar screens done with C. elegans with the same molecules (SC, JK, PJR, unpublished results), the hit rate with D. dipsaci is significantly lower, and many drug plates exhibit no activity (Figure 1A). Some plates have fully reproducible hits (Figure 1B,C), while others vary in activity (Figure 1C). Relative to other species screened (not shown), D. dipsaci shows less variability in its response to compounds. Regardless, replicates are considered necessary in the identification of reproducible hits.
Nematicides vary in their ability to immobilize Ditylenchus dipsaci
Of the seven characterized nematicides tested against D. dipsaci, only Fluopyram exhibits robust activity in the assay described herein (Figure 1C). This is consistent with previous work showing that D. dipsaci is tolerant of nematicides13,14. Fluopyram inhibits complex II of the electron transport chain in a nematode-selective manner and is a commercial nematicide used to control a variety of PPNs, including Rotylenchulus reniformis and Meloidogyne incognita4,21,22. Fluopyram and one of the nematicides that lacked activity at the concentration tested (Oxamyl)23 were investigated in more detail through a dose-response analysis with D. dipsaci. Fluopyram induces an apparent dose-dependent effect on D. dipsaci mobility with an EC50 of 9.3 μM (with a 95% confidence interval between 8.2 to 10.5 μM) (Figure 2A,B). This result is expected based on published in vitro results from Storelli et al., 202013. Oxamyl has no significant effect on mobility up to a concentration of 120 µM (p = 0.3632, unpaired t-test) (Figure 2C,D).
Sodium hydroxide improves assay sensitivity
In contrast to the near-continuous swimming activity of C. elegans in liquid culture18, D. dipsaci animals are dramatically less mobile. This is not uncommon among parasitic nematodes in culture16. To help distinguish 'resting' worms from sick worms, 40 mM of NaOH is used at the assay endpoint to stimulate movement in those individuals who are capable (Figure 3)19,20. This technique allows for the clear identification of small molecules that immobilize worms, given that all worms in negative control wells move and yield an exceptionally low background of false positives in the screen.
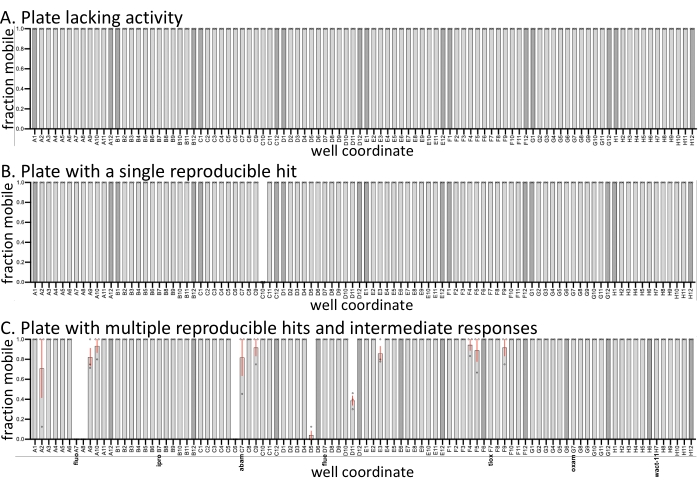
Figure 1: Examples of screen output. Three biological replicates (open circles) with D. dipsaci were performed against the small molecules (60 µM) in each of the three plates shown. All three plates have 0.6% of DMSO solvent-only controls (darker bars). Except where otherwise noted with solvent controls or known nematicides, the wells of the three plates contain relatively uncharacterized drug-like compounds purchased from vendors (see Burns et al., 2015)18. (A) A 96 well plate from the small molecule library lacks any molecule with observable bioactivity against D. dipsaci. (B) A 96 well plate from the small molecule library has a single molecule that reproducibly disrupts D. dipsaci mobility. (C) A 96 well drug plate containing the characterized nematicides fluopyram (fluo), iprodione (ipro), abamectin (abam), fluensulfone (flue), tioxazafen (tiox), oxamyl (oxam), and wact-11. The error bars represent the standard error of the mean. Please click here to view a larger version of this figure.
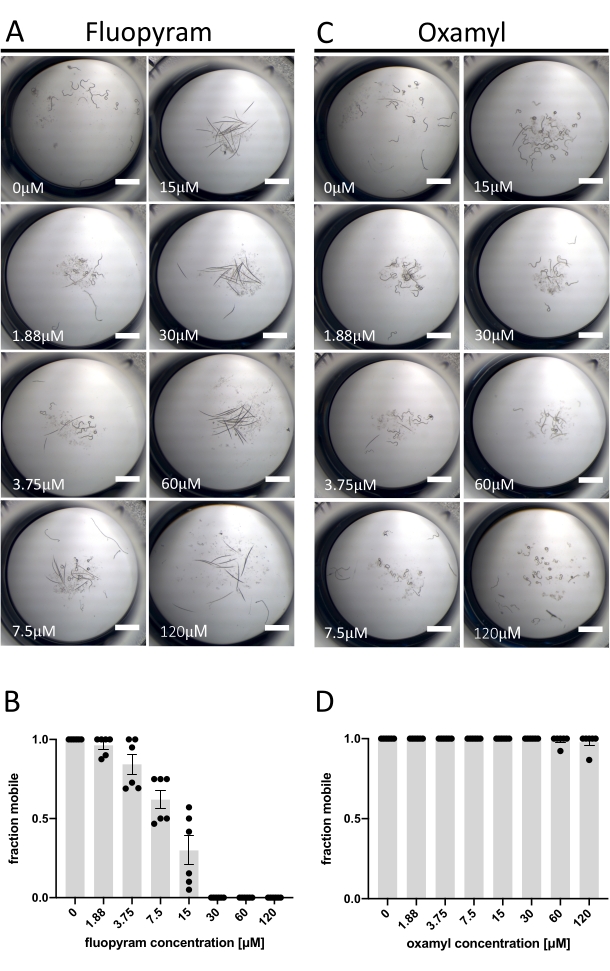
Figure 2: Example of positive and negative results using mobility assay. (A) Examples of the terminal phenotypes after the exposure of D. dipsaci to increasing concentrations of fluopyram after 5 days before addition of NaOH. (B) A dose-response analysis of the movement of D. dipsaci after 5 days of exposure to the indicated concentrations of fluopyram. (C,D) The same as A,B, except for oxamyl. Each graph shows trials done on two separate days with three replicates each day. The y-axis of each graph indicates the fraction mobile of the worms in each well calculated as the number of animals moving relative to the total number of animals in the well. The error bars on both graphs represent the standard error of the mean. The scale bar represents 1 mm. Please click here to view a larger version of this figure.
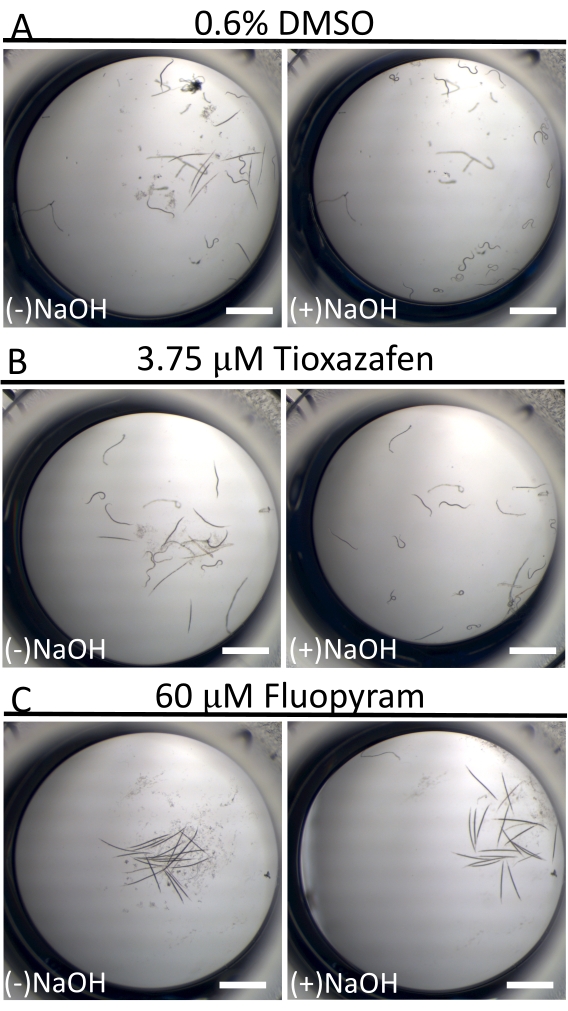
Figure 3: Mobility of D. dipsaci after adding 40 mM of NaOH. The effect of NaOH addition to D. dipsaci samples in the presence of solvent-only control (A), tioxazafen (B), or fluopyram (C) after 5 days of co-incubation. The scale bar represents 1 mm. Please click here to view a larger version of this figure.
Discussion
Critical steps
Despite the protocol's simplicity, there are critical steps in the protocol that deserve additional attention to maximize the likelihood of success. First, overbleaching the seeds can disrupt their growth. Therefore, limiting the seeds' time in the bleaching solution to 20 minutes or less is essential. Second, as previously noted by Storelli et al., the apparent health of the nematodes decreases over time when stored at 4 °C16. Using the nematodes soon after their collection provides additional confidence that optimal screening conditions can be achieved. Should longer-term storage be necessary, ensure that the tube's lid is not tight to allow oxygen exchange. Third, ensuring that the peas grow on the NA plates for the suggested time enables the experimenter to judge which seeds are contaminated. Fourth, overgrowing the peas on the GA plates before adding the nematodes will weaken infection and reduce the nematode yield. Finally, many factors that are difficult to control can impact screening results. Therefore, it is essential to perform multiple independent replicate screens on different days and ideally with PPNs collected from different culture plates to ensure the reproducibility of results.
Limitations of method
A limitation to the protocol is that it fails to synchronize the developmental stage of the collected worms, which range from juveniles to adults. Hence, strong hits revealed by any screen are likely effective at multiple stages. However, the protocol increases the risk of overlooking effective stage-specific hits. A second consideration is that in vitro screening should be considered the first step in a nematicide-discovery pipeline; soil-based assays are an excellent addition to a pipeline to test the translatability of the hits.
Significance and application of the protocol
The protocols described herein are simple and easily replicated. Furthermore, this protocol has been successfully applied to other PPNs in the lab, including Pratylenchus penetrans, by making only slight modifications. Developing new and safe PPN control measures is essential to ensure global food security. This is especially true for species like D. dipsaci that are generally tolerant to a wide variety of currently acceptable chemical nematicides13,14. Hence, the protocols outlined here have the potential to make an important contribution to human health on a global scale.
Disclosures
The authors declare no conflicts of interest.
Acknowledgements
The authors acknowledge Dr. Qing Yu (Agriculture and Agri-Food Canada) for providing the Ditylenchus dipsaci culture and for advice on culture methods; Dr. Benjamin Mimee (Agriculture and Agri-Food Canada) and Nathalie Dauphinais (Agriculture and Agri-Food Canada) for advice on in vitro culture of plant-parasitic nematodes; Dr. Andrew Burns and Sean Harrington for helpful suggestions on the project and the manuscript. JK is an NSERC Alexander Graham Bell Canada Graduate Scholar. PJR is supported by a CIHR project grant (313296). PJR is a Canada Research Chair (Tier 1) in Chemical Genetics.
Materials
| Name | Company | Catalog Number | Comments |
| 15 mL tube | Sarstedt | 62.554.205 | |
| 2 forceps | Almedic | 7747-A10-108 | |
| 2L beaker | Pyrex | CLS10002L | |
| 50mL beaker | Pyrex | CLS100050 | |
| 96 well plate | Sarstedt | 83.3924 | |
| aluminium foil | Alcan Plus | ||
| bacteriological agar | BioShop | AGR001.5 | |
| coffee filter | No name brand | 716 | |
| commercial bleach 6% | Lavo Pro 6 | DIN102358107 | |
| disposable petri dishes (10cm x 1.5cm) | Fisherbrand | FB0875712 | |
| disposable petri dishes (10cm x 2.5cm) | Sigma-Aldrich | Z358762 | |
| dissecting scope | Leica | Leica MZ75 | |
| DMSO | Sigma-Aldrich | 472301-500ML | |
| Funnel | VWR | 414004-270 | |
| Gamborg B5 | Sigma-Aldrich | G5893-10L | |
| glass petri dish | VWR | 75845-546 | |
| glass slide | MAGNA | 60-1200 | |
| Lint-Free Blotting Paper | V&P Scientific | VP 522-100 | |
| Low retention pipette tips (200uL) | LABFORCE | 1159M44 | |
| mutlichannel pipette | Eppendorf research plus | 3125000036 | |
| NaOH | Sigma-Aldrich | S8045-500G | |
| nutrient agar | Sigma-Aldrich | 70148-100G | |
| parafilm | Bemis | PM-996 | |
| pea seeds | Ontario Seed Company | D-1995-250G | |
| pin cleaning solutions | V&P Scientific | VP110A | |
| pinner | V&P Scientific | VP381N | |
| pinner rinse trays | V&P Scientific | VP 421 | |
| reagent reservoir with lid for multichannel pipettes | Sigma-Aldrich | BR703459 | |
| shaking incubator | New Brunswick Scientific | I26 | |
| sterile surgical blade | MAGNA | sb21-100-a | |
| stir bar | Fisherbrand | 2109 - 1451359 | |
| sucrose | BioShop | SUC507.1 |
References
- Sasser, J. N., Freckman, D. W. A. world perspective on nematology: the role of the society. Vistas on nematology. , Hyattsville: Society of Nematologists Inc. 7-14 (1987).
- Abad, P., et al. Genome sequence of the metazoan plant parasitic nematode Meloidogyne incognita. Nature Biotechnology. 26 (8), 909-915 (2008).
- Auwal, M., Hassan, T. H. P., Hongli, S., Jingwu, Z. Nematodes threats to global food security. Acta Agriculturae Scandinavica, Section B - Soil & Plant Science. 63 (5), 420-425 (2013).
- Chen, Z. X., Chen, S. Y., Dickson, D. W. Nematicides: Past and present uses. Nematology Vol 2, Nematode Management and Utilization. , CABI Publishing. 1179-1200 (2004).
- Desaeger, J., Wram, C., Zasada, I. New reduced-risk agricultural nematicides - and review. Journal of nematology. 52, (2020).
- Donely, N. The USA lags behind other agricultural nations in banning harmful pesticides. Environmental Health. 18 (1), 44(2019).
- Polansek, T. Monsanto halts launch of chemical after users complain of rashes. Reuters. , (2017).
- Sturhan, D., Brzeski, M. W. Stem and bulb nematodes, Ditylenchus spp. Manual of agricultural nematology. , 423-464 (1991).
- The European and Mediterranean Plant Protection Organization (EPPO). Data Sheets on Quarantine Pests; Ditylenchus dipsaci. Prepared by CABI and EPPO for the EU under Contract 90/399003. The European and Mediterranean Plant Protection Organization (EPPO). , (2021).
- Quebec (Canada), Agriculture, Pêcherie et Alimentation Quebec. Réseau d'avertissements phytosanitaires (RAP). Avertissement Carotte, Céleri, Laitue, Oignon, Poireau [Warning carrot celery, lettuce, onion, leek]. Quebec (Canada), Agriculture, Pêcherie et Alimentation Quebec. , (2011).
- Abawi, G. S., Moktan, K. Bloat nematode problem on garlic: symptoms, distribution, and management guidelines. Department of Plant Pathology and Plant-microbe Biology, Geneva. , Cornell University. (2010).
- Taylor, R. Chapter 7. Nematodes and other worms. , Academic Press. 143-234 (2019).
- Storelli, A., Keiser, A., Eder, R., Jenni, S., Kiewnick, S. Evaluation of fluopyram for the control of Ditylenchus dipsaci in sugar beet. Journal of nematology. 52, 1-10 (2020).
- Oka, Y. Nematicidal activity of fluensulfone against some migratory nematodes under laboratory conditions. Pest Management Science. 70, 1850(2014).
- Poirier, S., Dauphinais, N., Van Der Heyden, H., Véronneau, P. Y., Bélair, G., Gravel, V., Mimee, B. Host Range and Genetic Characterization of Ditylenchus dipsaci Populations from Eastern Canada. Plant disease. 103, 456-460 (2019).
- Storelli, A., Kiewnick, S., Daub, M., et al. Virulence and pathogenicity of four Ditylenchus dipsaci populations on sugar beet. Eur J Plant Pathol. 161, 63-71 (2021).
- Kühnhold, V., Kiewnick, S., Sikora, R. A. Development of an in vivo bioassay to identify sugar beet resistance to the stem nematode Ditylenchus dipsaci. Nematology. 8 (5), 641-645 (2006).
- Burns, A., Luciani, G., Musso, G. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat Communications. 6, (2015).
- Chen, S. Y., Dickson, D. W. A Technique for Determining Live Second-stage Juveniles of Heterodera glycines. Journal of nematology. 32, 117-121 (2000).
- Xiang, N., Lawrence, K. S. Optimization of In Vitro Techniques for Distinguishing between Live and Dead Second Stage Juveniles of Heterodera glycinesand Meloidogyne incognita. PLoS ONE. 11 (5), (2016).
- US EPA. Fluopyram. US EPA. , OFFICE OF CHEMICAL SAFETY AND POLLUTION PREVENTION (2016).
- US EPA. Fluopyram. US EPA. , OFFICE OF CHEMICAL SAFETY AND POLLUTION PREVENTION (2020).
- US EPA. Oxamyl Facts. Prevention, Pesticides and Toxic Substances. US EPA. , (2000).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved