A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
High-Throughput Screening of Microbial Isolates with Impact on Caenorhabditis elegans Health
* These authors contributed equally
In This Article
Summary
Gut microbes may positively or negatively impact the health of their host via specific or conserved mechanisms. Caenorhabditis elegans is a convenient platform to screen for such microbes. The present protocol describes high-throughput screening of 48 bacterial isolates for impact on nematode stress resistance, used as a proxy for worm health.
Abstract
With its small size, short lifespan, and easy genetics, Caenorhabditis elegans offers a convenient platform to study the impact of microbial isolates on host physiology. It also fluoresces in blue when dying, providing a convenient means of pinpointing death. This property has been exploited to develop high-throughput label-free C. elegans survival assays (LFASS). These involve time-lapse fluorescence recording of worm populations set in multiwell plates, from which population median time of death can be derived. The present study adopts the LFASS approach to screen multiple microbial isolates at once for the effects on C. elegans susceptibility to severe heat and oxidative stresses. Such microbial screening pipeline, which can notably be used to prescreen probiotics, using severe stress resistance as a proxy for host health is reported here. The protocol describes how to grow both C. elegans gut microbiota isolate collections and synchronous worm populations in multiwell arrays before combining them for the assays. The example provided covers the testing of 47 bacterial isolates and one control strain on two worm strains, in two stress assays in parallel. However, the approach pipeline is readily scalable and applicable to the screening of many other modalities. Thus, it provides a versatile setup to rapidly survey a multiparametric landscape of biological and biochemical conditions that impact C. elegans health.
Introduction
The human body harbors an estimated 10-100 trillion live microbial cells (bacteria, archaea fungi), which are primarily found in the gut, skin, and mucosal environments1. In a healthy state, these provide benefits to their host, including vitamin production, maturation of the immune system, stimulation of innate and adaptive immune responses to pathogens, regulation of fat metabolism, modulation of stress responses, and more, with an impact on growth and development, disease onset, and ageing2,3,4,5. The gut microbiota also evolves considerably throughout life. The most drastic evolution occurs during infancy and early childhood6, but significant changes also occur with age, including a decrease in Bifidobacterium abundance and an increase in Clostridium, Lactobacillus, Enterobacteriaceae, and Enterococcus species7. Lifestyle can further alter gut microbial composition leading to dysbiosis (loss of beneficial bacteria, overgrowth of opportunistic bacteria), resulting in various pathologies such as inflammatory bowel disease, diabetes, and obesity5, but also contributing to Alzheimer's and Parkinson's diseases8,9,10,11.
This realization has critically contributed to refining the concept of the gut-brain axis (GBA), where interactions between gut physiology (now including the microbes within it) and the nervous system are considered the main regulator of animal metabolism and physiological functions12. However, the precise role of microbiota in gut-brain signaling and the associated mechanisms of action are far from being fully understood13. With gut microbiota being a key determinant of healthy aging, how bacteria modulate the aging process has become a subject of intense research and controversy6,14,15.
With the demonstration that the roundworm Caenorhabditis elegans hosts a bonafide gut microbiota dominated-as in other species-by Bacteroidetes, Firmicutes, and Actinobacteria16,17,18,19,20, its rapid rise as an experimental platform to study host-gut commensal interactions21,22,23,24,25,26 has significantly expanded our investigative arsenal26,27,28,29. In particular, high-throughput experimental approaches available for C. elegans to study gene-diet, gene-drug, gene-pathogen, etc. interactions, can be adapted to rapidly explore how bacterial isolates and cocktails impact C. elegans health and aging.
The present protocol describes an experimental pipeline to screen at once arrays of bacterial isolates or mixtures set in multiwell plates for effects on C. elegans stress resistance as a proxy for health, which can be used to identify probiotics. It details how to grow large worm populations and handle bacterial arrays in 96- and 384-well plate formats before processing worms for automated stress resistance analysis using a fluorescence plate reader (Figure 1). The approach is based on label-free automated survival assays (LFASS)30 that exploit the phenomenon of death fluorescence31, whereby dying worms produce a burst of blue fluorescence that can be used to pinpoint the time of death. Blue fluorescence is emitted by glucosyl esters of anthranilic acid stored in C. elegans gut granules (a type of lysosome-related organelle), which burst when a necrotic cascade is triggered in the worm gut upon death31.
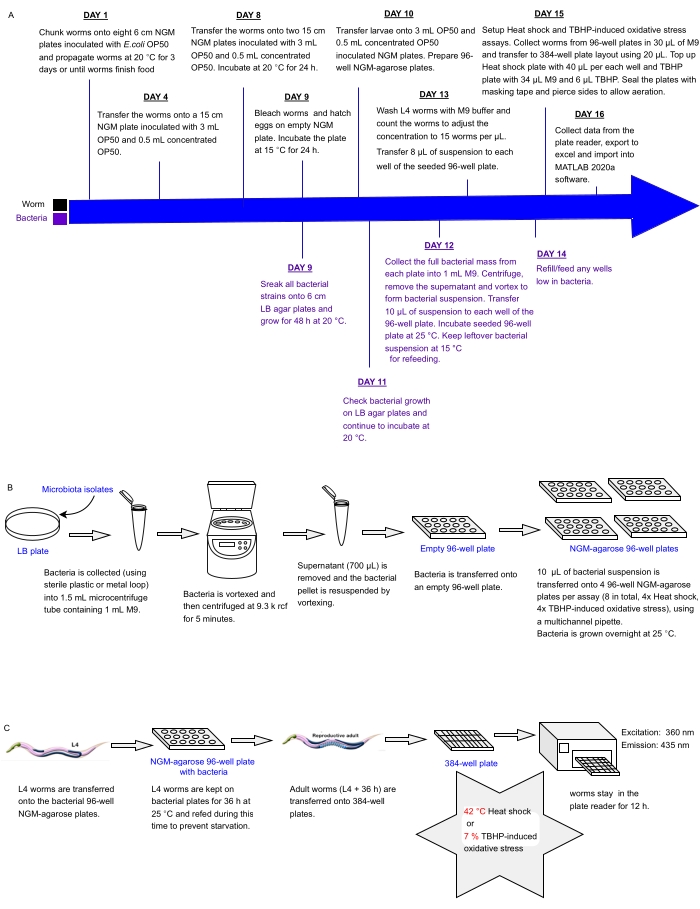
Figure 1: Experimental workflow for high-throughput screening of bacterial isolates with impact on C. elegans resistance to stress. (A) Timeline for worm and bacterial maintenance and assay setup. (B) 96-well bacterial plate array setup and handling. (C) 384-well worm plate setup. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Protocol
The two C. elegans strains used in parallel for the present study were Bristol N2 wild type and HT1890: daf-16(mgDf50), which grow at similar rates. However, the protocol can be replicated with any combination of two strains that have similar growth rates. Note that, when testing other strains in parallel (for instance, wild type and slow-growing daf-2 mutants), differing growth rates must be considered, and accordingly, the protocol needs to be adjusted. The timescales and quantities of worms and bacteria in the following protocol are optimized for parallel testing of 48 bacterial isolates on two worm strains in two LFASS assays in tetraplicates. Adjustments will be needed if more conditions are to be tested in parallel. Escherichia coli OP50 bacteria strain was obtained from the Caenorhabditis Genetics Center (CGC), University of Minnesota. The 48 bacterial isolates were obtained from the Schulenburg lab and maintained on LB agar.
1. C. elegans culturing on OP50 (Days 1 - 8)
NOTE: The current approach aims to grow C. elegans hermaphrodites on a solid medium at all stages and avoids unnecessary dietary changes (i.e., using alternate faster-growing E. coli strains such as NA22 or richer growth media such as egg plates) to remain as close as possible to the standard growth conditions32,33 that are still widely used. Worm growth temperature (here set at 15 °C) depends on the C. elegans strain(s) used and may need adjusting (for instance, to avoid or trigger the expression of a temperature-sensitive phenotype or biomarker). For information on worm husbandry, please see reference33.
- Prepare eight 6 cm diameter NGM plates (10 mL of nematode growth media agar, NGM, Supplementary File 1)32,33 per worm strain and let them dry for 1 day at room temperature.
- Prepare a saturated liquid culture of E. coli OP50 bacteria by seeding a single bacterial clone from a freshly grown Lysogeny Broth agar (LB agar, Supplementary File 1) plate in 25 mL of OP50 medium (Supplementary File 1) in a 50 mL conical tube. Grow the culture overnight at 37 °C in a shaker incubator.
- Inoculate the eight 6 cm NGM plates per strain with 100 µL of saturated liquid culture of E. coli OP50 per plate and keep the plates at 20 °C for 2 days before use.
- Using a scalpel, cut and transfer a square agar chunk of 0.5 cm with worms from a recently starved NGM plate onto each of the eight inoculated 6 cm NGM plates and incubate these plates at 20 °C for 3 days (or until the worms finish the food).
- Prepare five 15 cm NGM plates per worm strain (30 mL of NGM medium per plate) and inoculate with 3 mL of OP50. Let the plates dry before incubating at 37 °C overnight. Keep the plates at 20 °C until use in later steps.
- Using a P-1000 pipette, add up to 3 mL of sterile M9 buffer (Supplementary File 1) to the 6 cm NGM plates (Step 1.1.) to resuspend the worms, and collect the worm solution from all eight plates per strain in a single 15 mL conical tube.
- Centrifuge at 142 x g for 2 min at 4 °C. Carefully remove the supernatant using a P-5000 pipette or a water pump equipped with a sterile Pasteur pipette or tip. Add 10 mL of sterile M9 buffer to wash the worm pellet. Repeat 2x.
- Remove the supernatant (as much as possible) and transfer the worms onto a 15 cm OP50 inoculated NGM plate (Step 1.5.) using a pipette. Add 0.5 mL of concentrated OP50 culture.
- To make concentrated OP50, inoculate each of four 1 L bottles of LB with 2 mL of OP50 starter culture (prepared in Step 1.2.), and grow in a shaking incubator for 6 h at 37 °C and 160 x g. Pellet the bacteria at 3057 x g and 20 °C for 15 min. Discard the supernatant, resuspend the bacterial pellets with 6 mL of OP50 medium, and collect in a sterile 50 mL conical tube.
NOTE: Bacteria can be stored at 4 °C for up to 1 week.
- Grow each worm strain on a 15 cm diameter NGM plate for 3-4 days at 15 °C by re-feeding the worms with 0.5 mL of concentrated OP50 daily.
- Once the worms have nearly finished the food, collect and wash in M9 buffer (Step 1.6.1.), transfer each worm strain culture to two 15 cm NGM plates (Step 1.5.), and propagate worms at 20 °C until ~95% of the population are gravid adults (it will take about 24 h for wild type Bristol N2).
NOTE: Gravid adults are characterized by the presence of eggs within the worm, and the ideal plate should also have an abundance of unhatched eggs laid on the plate without too many larvae33.
- Once the worms have nearly finished the food, collect and wash in M9 buffer (Step 1.6.1.), transfer each worm strain culture to two 15 cm NGM plates (Step 1.5.), and propagate worms at 20 °C until ~95% of the population are gravid adults (it will take about 24 h for wild type Bristol N2).
2. Maintenance of gut microbiota isolate collections (Day 9)
- Streak the 48 bacterial isolates on individual 6 cm LB agar plates and grow for 48 h at 20 °C.
NOTE: Bacteria can be grown at 25 °C for 24-36 h if needed sooner, but the longer 20 °C growth allows for spotting potential contaminants. - Synchronize a large number of C. elegans.
- Bleach gravid adult worms by following the standard egg preparation method33 and transfer the eggs onto two unseeded 15 cm NGM plates for 24 h at 15 °C to allow for all the L1 larvae to hatch and grow synchronously in the subsequent steps.
CAUTION: Be careful while handling bleach solutions.
- Bleach gravid adult worms by following the standard egg preparation method33 and transfer the eggs onto two unseeded 15 cm NGM plates for 24 h at 15 °C to allow for all the L1 larvae to hatch and grow synchronously in the subsequent steps.
3. Growing large C. elegans cultures (Day 10)
- Once hatched, collect the L1 larvae (from Step 2.2.1.) in 3-4 mL of M9 in a clean conical 15 mL tube. Pipette four 10 µL drops of worm solution onto a slide or a plate and count the number of worms in each drop under a stereomicroscope at 16x magnification. Determine the worm concentration of the solution by averaging the number of larvae from all the drops of worm solution. Multiply this value by the volume left and estimate the total worm count for each strain.
NOTE: 46,000-50,000 L1 larvae per strain are required at this stage to later fill a 384-well plate or two half-plates.- For each strain, transfer all the L1 larvae onto two 15 cm NGM plates (23,000-25,000 L1 per plate) previously inoculated with 3 mL of OP50 (Step 1.5.) and re-seeded with 0.5 mL of concentrated OP50.
- Incubate at 15 °C, topping up with 0.5 mL of concentrated OP50 daily as needed until the worms reach the L4 stage.
NOTE: The L4 stage is characterized by a slightly darker intestine and a half-disk or crescent-shaped white patch where the vulva will eventually form32,33. - Prepare 96-well NGM-agarose plates following the steps below.
- Prepare eight 96-well NGM-agarose plates by filling each well with 125 µL of NGM-agarose (four plates per assay).
NOTE: It is recommended to plate some extra plates if some get contaminated in the subsequent steps. Two plate readers will be required to run assays in parallel, but they may also be run successively starting with the heat stress assay as it can be run for as little as 6 h. For these plates, the <4% ash agar is substituted with agarose (see Table of Materials), enabling slower and more even drying across the NGM plugs and reducing worm burrowing for better recovery. - Ensure that the wells are filled evenly and bubble-free. Use a heat block set at 70 °C (with slow heat transfer through the plastic of the multiwell plate, the NGM-agarose may only heat up to about 55-60 °C) to keep the mixture from solidifying during the process. To remove bubbles within wells, use a sterile flame-heated needle.
- Allow the 96-well plates to set at room temperature in a sterile environment before inverting (lid down to prevent condensation) and store at 4 °C in a clean box until needed.
- Prepare eight 96-well NGM-agarose plates by filling each well with 125 µL of NGM-agarose (four plates per assay).
- On Day 11, check on the worms from Step 3.2., ensuring no contaminations have appeared and the worms are still replete.
- On Day 12, check on the worms from Step 3.1., ensuring no contaminations have appeared and the worms are still replete. Also, check the worms' developmental stage.
NOTE: The sex/strain and the developmental stage of the worm, such as L4 or L4 + 24 h worms used, are dependent on the treatments the worms are subjected to. Here, wild-type hermaphrodites were exposed to bacterial isolates from L4 for 36 h.
4. Preparing gut microbiota isolate collections for re-feeding worms
- Monitor bacterial growth on the LB agar plates from Step 2.1. and continue to incubate at 20 °C.
NOTE: While it is not ideal, in case some clones do not grow or reveal contaminations, bacteria may be re-streaked from clean stocks onto 6 cm LB plates and grown at 25-28 °C for 24 h to be ready for the experiment. - Define a 96-well array layout for the bacterial collection being tested, facilitating systematic plate seeding and data analysis in the subsequent steps (Supplementary Table 1).
- Collect the bacterial mass from each 6 cm bacterial plate (Step 4.1.), and transfer it to a labeled 1.5 mL microcentrifuge tube containing 1 mL of M9 buffer. Perform this by using either a single-use 2 mm diameter sterile plastic loop or a 5 mm diameter metal loop. Sterilize the metal loop between bacterial strains by dipping in 100% ethanol, flaming, and cooling down for 5 s.
- Vortex the microcentrifuge tubes until the bacterial pellets are fully resuspended (depending on the bacterial strain, this may take ~1-10 s).
- Spin down at 9,300 x g for 5 min at room temperature, remove 700 µL of supernatant, and resuspend the bacterial pellet by vortexing.
- Transfer 200 µL of each bacterial suspension into a single well of an empty sterile 96-well plate according to the layout set out in Step 4.2.
- From this plate, inoculate eight 96-well NGM-agarose plates (prepared in Step 3.3.) with 10 µL of bacterial solution using a multichannel pipette and incubate with the lid on at 25 °C for 24 h. Do not seal the plates to allow for plate drying and bacterial aerobic growth and to avoid excess condensation.
- Seal the 96-well suspension plate prepared in Step 4.6. with clean adhesive sealing film (see Table of Materials), and store at 15 °C for up to 5 days. This will be used for worm re-feeding as needed.
5. LFASS heat shock and oxidative assay setup (Days 13 - 14)
- Looking at the plates from Step 3.5., assess the worms' developmental stage. Once >90% of worms have reached L434, collect the worms in up to 10 mL of sterile M9 solution in 15 mL conical tubes.
- Wash the worms extensively (at least 4x) by spinning down at 142 x g for 2 min at 4 °C, removing the supernatant, and adding 10 mL of fresh sterile M9 between each wash to get rid of OP50 bacteria. Resuspend the worm pellet in 10 mL of M9.
- Transfer 50 µL of worm solution into a low surface binding tube (see Table of Materials) containing 950 µL of M9. After gently mixing the tube contents to avoid worm sedimentation, quickly use a wetted low-bind pipette tip to transfer 3-4 separate 10 µL drops onto a glass slide or an NGM plate, and count worm numbers under a stereomicroscope (see Table of Materials) at 16x magnification. Average the counts from the 3-4 drops and determine the number of worms per microliter in the worm solution (see Step 3.1.).
- Adjust the worm concentration in the 10 mL tube to reach ~120 worms in 8 µL. If the solution prepared in Step 5.2. is not concentrated enough, spin the worms down and remove M9 accordingly to reach 120 worms per 8 µL.
- Transfer 8 µL of worm solution (~120 worms) into each of the wells of the eight 96-well NGM-agarose plates from Step 4.7., using a multichannel pipet or a repeat pipet. Ensure to use low retention tips to limit worm loss. It might also be necessary to cut the tip ends to allow for large adult worms to limit mechanical stress on adult worms.
NOTE: The assay requires a minimum of 30 live healthy worms to work reliably but works best with about 100 worms per well. - Incubate the worm and bacterium-seeded 96-well NGM-agarose plates at 25 °C for 36 h.
- Check the plates between 12-24 h, ensuring the worms remain replete throughout. If re-feeding is required, resuspend the bacteria within the 96-well bacterial array plate stored at 15 °C in Step 4.8., and add up to 10 µL of the corresponding bacterial solution to the 96-well NGM-agarose plates where worms are at risk of starvation before the end of the 36 h incubation period (starved worms will produce vastly differing results, so this is very important).
NOTE: The following steps need to be conducted on Day 15. Prior to starting the assay, it may be necessary to optimize the reading height. The optimal reading will be achieved 20-50 µm above the bottom of the well. This will be dependent on the model of the plate reader. Some offer the possibility of a Z-scan, while others allow manual height input. Set the optimal height at the level where the highest blue fluorescence (365 nm/430 nm) signal is detected. Some plate-readers may operate at a fixed height optimized for adherent cell assays and might not be ideal for LFASS assays. - After 36 h, dispense 30 µL of M9 into each well of the 96-well plate.
NOTE: For thermal stress assays, the plate reader needs to have reached the required temperature to perform the assay and may need to be turned on ahead of time. The current protocol uses 42 °C to maximize killing speed, but the approach applies to other temperatures above 30 °C. - Transfer worms (about 20 µL) to the 384-well plate according to set layouts, using low-retention tips (consider cutting off the end of the tips to allow large worms to reduce mechanical stress for adult worms).
NOTE: For the present study, two different plate reader settings are used for the two assays described here (thermal stress and oxidative stress), and thus samples intended for these two assays must not be plated in the same 384-well plate. - Ensure the plate readers are set up properly (Table 1).
- Top up the 384-well plates with more M9, aiming for a final volume of 60 µL per well. For thermal stress assay, add 40 µL of M9, and for t-BHP-induced oxidative stress, add 34 µl of M9 in 6 µl of t-BHP (see Table of Materials).
- Start the assay within 2 min of adding t-BHP (ideally, all worms must be exposed to t-BHP simultaneously, the assay time resolution being 2 min). If not possible, use a timer to estimate the time spent pipetting t-BHP before the start of the assay to allow for later adjustment of the median time of death.
- Close the plates with their transparent lid. Seal the edges of the 384-well plates with masking tape (taping over the plate and lid), ensuring that the tape does not go over the lid or under the plate. Slit the tape between lid and plate at intervals using a scalpel to allow air exchange while minimizing evaporation during the assay.
- Insert the plate into the plate reader (see Table of Materials) and start the run. Aim to excite at 365 nm and detect emission at 435 nm every 2 min for 6-12 h (Table 1).
NOTE: Typically, 6 h is enough for 42 °C heat stress assays and 8 h for 7% t-BHP oxidative stress assays.
6. Plate-reader data handling
- Save the raw fluorescence data from the plate-reader as comma- or tab-separated .txt, .csv, or .xls /.xlsx formats, and then convert to xls /.xlsx format. Depending on the data format, reorganize them to match the excel sheet layout needed for LFASS analysis. Follow the detailed instructions provided in reference30.
NOTE: While data can be analyzed manually, normalizing each time series and looking for the time when death fluorescence reaches the half maximum, automated analysis can be carried out in Matlab running the LFASS routine30. - Download and install Matlab (version 2014a or above) and the LFASS software package from https://github.com/ABA80/LFASS. Follow the guidelines and annotations provided within it.
NOTE: Figure 1C gives a brief description of the approach. Matlab is required to run the LFASS routine. Alternatively, the Matlab code may be translated into Oracle, except for the fitting function, which is proprietary. New smoothing and sigmoid functions can be rewritten to enable use in a fully open-source platform. - Between LFASS analyses, move the data and results to a new location as the LFASS analysis will process all files in the data folder and overwrite files in the Results folder.
7. Data inspection
- Open the excel file and label the rows according to the well position on the 384-well plate. Supplementary File 2 shows an example of the excel file of the raw fluorescence data generated for the heat shock assay. Use the well position on the 384-well plate to label the worm and bacterial strains.
- Ahead of Matlab analysis, visually inspect the data in excel, plotting fluorescence intensity over time for a representative well. Depending on the plate-reader used, data may be noisy but should display a clear peak. In particular:
- Determine a fluorescence value below which a peak would not be significantly different from noise (setting such a threshold in LFASS will speed up analysis by excluding empty wells).
- Note the earliest time point when fluorescence fluctuations dampen prior to rising (animals may thrash vigorously for up to 30 min, leading to fast fluctuating blue fluorescence readings).
NOTE: The peak fitting may be improved by excluding these early timepoints from the curve fitting window. - Note the time points between which minimal and maximal fluorescence values are expected to fall (look at several wells to identify these ranges) as they will be used for curve fitting.
- Check if the amplitudes of the fluorescence peaks vary significantly between the wells, normalize the data prior to further analysis using the following formula:
Normalized fluorescence welln (t) = (Fluorescence welln [t] - minimum fluorescence well [Dt]) / (maximum fluorescence well [Dt] - minimum fluorescence well [Dt])
where "n" is the current well number, "t" is the timepoint, and "Dt" is the series of time points for the assay.
8. LFASS data processing
NOTE: Details are provided at https://github.com/ABA80/LFASS and in the supplementary materials of Reference30.
- Create two subfolders within the LFASS folder, one for the data to be analyzed and one for results, for example, "my data" and "results".
- Copy the assay excel data file into the LFASS subfolder "my data" after data inspection.
- Launch MATLAB, navigate to the LFASS folder, type, and run fitfolder in the command window (Supplementary File 3). Then follow the on-screen instructions.
- After typing in "fitfolder", the system asks for the name of the folder in which the excel file is located, for example, 'my data'. Type in the name of your data folder (in this example, "my data").
- Follow the on-screen instructions, providing the various parameters requested.
- Enter "2" for the time interval between successive measurements in the current protocol (specifying this allows for the results to be expressed in minutes instead of timepoint units).
NOTE: The time interval can be modified to perform fluorescence measurements more or less frequently (to decrease or increase time resolution) and also depending on the plate reader capabilities (i.e., the time interval may need to be increased for plate readers that cannot perform fast enough measurements). Ensure always to match the experimental time interval with the LFASS routine. - Assign the upper tolerance threshold by typing "0.95" (this can be changed as needed to improve fit) and the lower tolerance threshold by typing "0.05" (this can be changed as needed to improve fit) to constrain the sigmoid fit.
NOTE: Other time parameters are based on user notes from the data inspection (Step 7.2.).
- Enter "2" for the time interval between successive measurements in the current protocol (specifying this allows for the results to be expressed in minutes instead of timepoint units).
- Choose whether to display or not fitted and smoothed curves by typing "y" for YES or "n" for NO when prompted. To visually inspect the converging fits, select the former.
NOTE: The latter is useful to visualize all smoothed data but is usually not selected because it generates too many pop-up graphs. Following this, Matlab will execute the LFASS routine, which may take 1-10 min if multiple excel files are being processed in one go. Pop-up windows with curves will appear according to the selection in Step 8.6. Supplementary File 4A shows an example of a fitted curve. - Choose whether to (1) analyze curves identified as noise or (2) refit poorly fitted curves with an [y/n] option. Type y to approve and n to reject.
NOTE: Approving refitting is recommended, especially if there are many poorly fitted or unfitted curves. This will allow the user to provide tailored curve fitting parameters for each curve as they appear on the screen and only ask for earlier and later boundaries for the sigmoid fit. It can be attempted as many times as required. - Once the data is analyzed, close Matlab and open the LFASS folder.
- Click on the LFASS subfolder My results, as the result files are saved automatically in the Results folder as .txt.
NOTE: Matlab generates three .txt files: "Batch-fitted.txt", "Batch and noise-fitted.txt", and "Refitted.txt". The former two are saved as a precaution in case of a computer crash or user error during the refitting. The file containing the most accurate complete analysis is "Refitted.txt". - Open the file Refitted.txt with Microsoft Excel and save as .xls for further processing. Supplementary File 4B shows an example of such a result file.
NOTE: For each well (organized in rows), three values are provided in the columns that give estimates of the median time of death of the worm population: "Raw": reports the time intersecting at the half-maximum of the experimental data peak; "Batch-fitted": reports the time to intersect at the half-maximum of the batch-fitted curve; "Refitted": reports the time to intersect at the half-maximum of the refitted curve. - Save the file in .xls format as a copy in a safe location. Failing to do this will run the risk of the files being overwritten during the next run of the LFASS routine.
NOTE: The results can then be further processed for graphing or statistical analysis.
Access restricted. Please log in or start a trial to view this content.
Results
LFASS assays provide robust, high-throughput, and rapid screening of multiple test conditions at once, such as screening numerous genetic and microbiota parameters that contribute to stress resistance and aging. It only takes 2-3 weeks for the experiment to acquire an extensive dataset of multiple test conditions. L4 + 36 h adult wild-type worm populations were exposed to 42 °C thermal stress and 7% t-BHP-induced oxidative stress after a 36 h culture on 48 gut microbial isolates for 36 h. The assay was perf...
Access restricted. Please log in or start a trial to view this content.
Discussion
C. elegans offers many advantages for rapidly screening multiple experimental parameters at once, owing to its small size, transparency, fast development, short lifespan, inexpensiveness, and ease of handling. Its considerably simpler genome, body plan, nervous system, gut, and microbiome, yet complex and similar enough to humans, make it a powerful preclinical model, where mechanistic insight can be gained while testing for bioactive efficacy or toxicity. As interest is growing in developing microbial intervent...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank the CGC Minnesota (Madison, USA, NIH - P40 OD010440) for providing worm strains and OP50 and Pr. Hinrich Schulenburg (CAU, Kiel, Germany) for providing all the environmental microbial isolates depicted here. This work was funded by a UKRI-BBSRC grant to AB (BB/S017127/1). JM is funded by a Lancaster University FHM PhD scholarship.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 10 cm diameter plates (Non-vented) | Fisher Scientific | 10720052 | Venting is not necessary for bacterial cultures |
| 15 cm diameter plates (Vented) | Fisher Scientific | 168381 | |
| 384-well black, transparent flat bottom plates | Corning | 3712 or 3762 | Not essential to be sterile for fast stress assays |
| 6 cm diameter plates (Vented) | Fisher Scientific | 150288 | Venting is necessary for worm cultures to avoid hypoxia |
| 96-well transparent plates (Biolite) | Thermo | 130188 | |
| Agar (<4% ash) | Sigma-Aldrich | 102218041 | Good quality agar is important for the structural integrity of the culture media, to avoid worm burrowing |
| Agarose | Fisher Scientific | BP1356 | |
| Avanti Centrifuge J-26 XP | Beckman coulter | ||
| Bleach | Honeywell | 425044 | |
| Calcium chloride | Sigma-Aldrich | C5080 | |
| Centrifuge 5415 R | Eppendorf | ||
| Centrifuge 5810 R | Eppendorf | ||
| Cholesterol | Sigma-Aldrich | C8667 | |
| LB agar | Difco | 240110 | |
| LB broth | Invitrogen | 12795084 | |
| LoBind tips | VWR | 732-1488 | Lo-bind reduce worm loss during transfers |
| LoBind tubes | Eppendorf | 22431081 | |
| Magnesium sulfate | Fisher Scientific | M/1100/53 | |
| Plate reader- infinite M nano+ | Tecan | Monochromator setup enables fluorescence tuning but adequate filter-based setups may be used | |
| Plate reader- Spark | Tecan | ||
| Potassium phosphate monobasic | Honeywell | P0662 | |
| Sodium chloride | Sigma-Aldrich | S/3160/63 | |
| Stereomicroscope setup with transillumination base | Leica | MZ6, or M80 | Magnification from 0.6-0.8x up to 40-60x is necessary, as is a good quality transillumination base with a deformable, titable or slidable mirror to adjust contrast |
| t-BHP (tert-Butyl hydroperoxide) | Sigma-Aldrich | 458139 | |
| Transparent adhesive seals Nunc | Fisher Scientific | 101706871 | It is important that it is transparent and that it can tolerate the temperatures involved in the assays. |
| Tryptophan | Sigma-Aldrich | 1278-7099 | |
| Yeast extract | Fisher Scientific | BP1422 |
References
- Krishna, S., et al. Integrating microbiome network: establishing linkages between plants, microbes and human health. The Open Microbiology Journal. 13, 330-342 (2019).
- Amon, P., Sanderson, I. What is the microbiome. Archives of Disease in Childhood - Education & Practice Edition. 102 (5), 257-260 (2017).
- Belkaid, Y., Harrison, O. J. Homeostatic immunity and the microbiota. Immunity. 46 (4), 562-576 (2017).
- Cabreiro, F., Gems, D. Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Molecular Medicine. 5 (9), 1300-1310 (2013).
- Vaga, S., et al. Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Scientific Reports. 10 (1), 14977(2020).
- Nagpal, R., et al. Gut microbiome and aging: Physiological and mechanistic insights. Nutrition and Healthy Aging. 4 (4), 267-285 (2018).
- Mitsuoka, T. Establishment of intestinal bacteriology. Biosci Microbiota Food Health. 33 (3), 99-116 (2014).
- Bonfili, L., et al. Microbiota modulation as preventative and therapeutic approach in Alzheimer's disease. The FEBS Journal. 288 (9), 2836-2855 (2021).
- Vendrik, K. E. W., et al. Fecal microbiota transplantation in neurological disorders. Frontiers in Cellular and Infection Microbiology. 10, 98(2020).
- Wang, Q., et al. The role of gut dysbiosis in Parkinson's disease: mechanistic insights and therapeutic options. Brain. 144 (9), 2571-2593 (2021).
- Zhu, X., et al. The relationship between the gut microbiome and neurodegenerative diseases. Neuroscience Bulletin. 37 (10), 1510-1522 (2021).
- Miller, I. The gut-brain axis: historical reflections. Microbial Ecology in Health and Disease. 29 (1), 1542921(2018).
- Foster, J. A., Rinaman, L., Cryan, J. F. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiology of Stress. 7, 124-136 (2017).
- Coman, V., Vodnar, D. C. Gut microbiota and old age: Modulating factors and interventions for healthy longevity. Experimental Gerontology. 141, 111095(2020).
- Conway, J., Duggal, N. A. Ageing of the gut microbiome: Potential influences on immune senescence and inflammageing. Ageing Research Reviews. 68, 101323(2021).
- Berg, M., et al. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. The ISME Journal. 10 (8), 1998-2009 (2016).
- Dirksen, P., et al. CeMbio - The Caenorhabditis elegans Microbiome Resource. G3: Genes, Genomes, Genetics. 10 (9), 3025-3039 (2020).
- Dirksen, P., et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biology. 14, 38(2016).
- Samuel, B. S., Rowedder, H., Braendle, C., Felix, M. A., Ruvkun, G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proceedings of the National Academy of Sciences of the United States of America. 113 (27), 3941-3949 (2016).
- Zimmermann, J., et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. The ISME Journal. 14 (1), 26-38 (2020).
- Dinic, M., et al. Host-commensal interaction promotes health and lifespan in Caenorhabditis elegans through the activation of HLH-30/TFEB-mediated autophagy. Aging. 13 (6), 8040-8054 (2021).
- Goya, M. E., et al. Probiotic Bacillus subtilis protects against alpha-Synuclein aggregation in C. elegans. Cell Reports. 30 (2), 367-380 (2020).
- Hacariz, O., Viau, C., Karimian, F., Xia, J. The symbiotic relationship between Caenorhabditis elegans and members of its microbiome contributes to worm fitness and lifespan extension. BMC Genomics. 22 (1), 364(2021).
- Shin, M. G., et al. Bacteria-derived metabolite, methylglyoxal, modulates the longevity of C. elegans through TORC2/SGK-1/DAF-16 signaling. Proceedings of the National Academy of Sciences of the United States of America. 117 (29), 17142-17150 (2020).
- Zhang, F., et al. Natural genetic variation drives microbiome selection in the Caenorhabditis elegans gut. Current Biology. 31 (12), 2603-2618 (2021).
- Zhang, F., et al. High-throughput assessment of changes in the Caenorhabditis elegans gut microbiome. Methods in Molecular Biology. 2144, 131-144 (2020).
- Chan, J. P., et al. Using bacterial transcriptomics to investigate targets of host-bacterial interactions in Caenorhabditis elegans. Scientific Reports. 9 (1), 5545(2019).
- Hartsough, L. A., et al. Optogenetic control of gut bacterial metabolism to promote longevity. Elife. 9, 56849(2020).
- Pryor, R., et al. Host-microbe-drug-nutrient screen identifies bacterial effectors of Metformin therapy. Cell. 178 (6), 1299-1312 (2019).
- Benedetto, A., et al. New label-free automated survival assays reveal unexpected stress resistance patterns during C. elegans aging. Aging Cell. 18 (5), 12998(2019).
- Coburn, C., et al. Anthranilate fluorescence marks a calcium-propagated necrotic wave that promotes organismal death in C. elegans. PLOS Biology. 11 (7), 1001613(2013).
- Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A., Ceron, J. Basic Caenorhabditis elegans methods: synchronization and observation. Journal of Visualized Experiments. (64), e4019(2012).
- Stiernagle, T. Maintenance of C. elegans. WormBook. , 1-11 (2006).
- Naomi, R., et al. Probiotics for Alzheimer's disease: a systematic review. Nutrients. 14 (1), 20(2021).
- Zheng, S. Y., et al. Potential roles of gut microbiota and microbial metabolites in Parkinson's disease. Ageing Research Reviews. 69, 101347(2021).
- Gill, M. S., Olsen, A., Sampayo, J. N., Lithgow, G. J. An automated high-throughput assay for survival of the nematode Caenorhabditis elegans. Free Radical Biology and Medicine. 35 (6), 558-565 (2003).
- Park, H. -E. H., Jung, Y., Lee, S. -J. V. Survival assays using Caenorhabditis elegans. Molecules and Cells. 40 (2), 90-99 (2017).
- Partridge, F. A., et al. An automated high-throughput system for phenotypic screening of chemical libraries on C. elegans and parasitic nematodes. International Journal for Parasitology: Drugs and Drug Resistance. 8 (1), 8-21 (2018).
- Rahman, M., et al. NemaLife chip: a micropillar-based microfluidic culture device optimized for aging studies in crawling C. elegans. Scientific Reports. 10 (1), 16190(2020).
- Stroustrup, N., et al. The Caenorhabditis elegans lifespan machine. Nature Methods. 10 (7), 665-670 (2013).
- Xian, B., et al. WormFarm: a quantitative control and measurement device toward automated Caenorhabditis elegans aging analysis. Aging Cell. 12 (3), 398-409 (2013).
- Brown, A. E., Schafer, W. R. Unrestrained worms bridled by the light. Nature Methods. 8 (2), 129-130 (2011).
- Churgin, M. A., et al. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife. 6, 26652(2017).
- Jushaj, A., et al. Optimized criteria for locomotion-based healthspan evaluation in C. elegans using the WorMotel system. PLoS One. 15 (3), 0229583(2020).
- Nambyiah, P., Brown, A. E. X. Quantitative behavioural phenotyping to investigate anaesthesia induced neurobehavioural impairment. Scientific Reports. 11 (1), 19398(2021).
- Squiban, B., Belougne, J., Ewbank, J., Zugasti, O. Quantitative and automated high-throughput genome-wide RNAi screens in C. elegans. Journal of Visualized Experiments. (60), e3448(2012).
- Zugasti, O., et al. Activation of a G protein-coupled receptor by its endogenous ligand triggers the innate immune response of Caenorhabditis elegans. Nature Immunology. 15 (9), 833-838 (2014).
- Zugasti, O., et al. A quantitative genome-wide RNAi screen in C. elegans for antifungal innate immunity genes. BMC Biology. 14, 35(2016).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved