A subscription to JoVE is required to view this content. Sign in or start your free trial.
Methods Article
Characterization of a Pathogenic Escherichia coli Strain Derived from Oreochromis spp. Farms Using Whole-Genome Sequencing
In This Article
Summary
The feasibility of whole-genome sequencing (WGS) strategies using benchtop instruments has simplified the genome interrogation of every microbe of public health relevance in a lab setting. A methodological adaptation of the workflow for bacterial WGS is described and a bioinformatics pipeline for analysis is also presented.
Abstract
Aquaculture is one of the fastest-growing food-producing sectors worldwide and tilapia (Oreochromis spp.) farming constitutes the major freshwater fish variety cultured. Because aquaculture practices are susceptible to microbial contamination derived from anthropogenic sources, extensive antibiotic usage is needed, leading to aquaculture systems becoming an important source of antibiotic-resistant and pathogenic bacteria of clinical relevance such as Escherichia coli (E. coli). Here, the antimicrobial resistance, virulence, and mobilome features of a pathogenic E. coli strain, recovered from inland farmed Oreochromis spp., were elucidated through whole-genome sequencing (WGS) and in silico analysis. Antimicrobial susceptibility testing (AST) and WGS were performed. Furthermore, phylogenetic group, serotype, multilocus sequence typing (MLST), acquired antimicrobial resistance, virulence, plasmid, and prophage content were determined using diverse available web tools. The E. coli isolate only exhibited intermediate susceptibility to ampicillin and was characterized as ONT:H21-B1-ST40 strain by WGS-based typing. Although only a single antimicrobial resistance-related gene was detected [mdf(A)], several virulence-associated genes (VAGs) from the atypical enteropathogenic E. coli (aEPEC) pathotype were identified. Additionally, the cargo of plasmid replicons from large plasmid groups and 18 prophage-associated regions were detected. In conclusion, the WGS characterization of an aEPEC isolate, recovered from a fish farm in Sinaloa, Mexico, allows insights into its pathogenic potential and the possible human health risk of consuming raw aquacultural products. It is necessary to exploit next-generation sequencing (NGS) techniques for studying environmental microorganisms and to adopt a one health framework to learn how health issues originate.
Introduction
Aquaculture is one of the fastest-growing food-producing sectors worldwide, and its production practices are intended to satisfy the rising food demand for human consumption. Global aquaculture production has tripled from 34 million tonnes (Mt) in 1997 to 112 Mt in 20171. The main species groups, contributing to nearly 75% of the production, were seaweed, carps, bivalves, catfish, and tilapia (Oreochromis spp.)1. However, the appearance of diseases caused by microbial entities is unavoidable because of intensive fish farming, leading to potential economic losses2.
Antibiotic usage in fish farming practices is well known for preventing and treating bacterial infections, the main limiting factor in productivity3,4. Nonetheless, residual antibiotics accumulate in aquaculture sediments and water, exerting selective pressure and modifying the fish-associated and the residing bacterial communities5,6,7,8. Consequently, the aquaculture environment serves as a reservoir for antimicrobial resistance genes (ARGs), and the further emergence and spread of antibiotic-resistant bacteria (ARB) in the surrounding milieu9. In addition to the bacterial pathogens commonly observed affecting fish farming practices, members of the Enterobacteriaceae family are often encountered, including human pathogen strains of Enterobacter spp., Escherichia coli, Klebsiella spp., and Salmonella spp.10. E. coli is the most common microorganism isolated from fish meal and water in fish farming11,12,13,14,15.
E. coli is a versatile gram-negative bacterium that inhabits the gastrointestinal tract of mammals and birds as a commensal member of their intestinal microbiota. However, E. coli possess a highly adaptive capacity to colonize and persist in different environmental niches, including soil, sediments, food, and water16. Because of the gene gain and loss through the horizontal gene transfer (HGT) phenomenon, E. coli has rapidly evolved into a well-adapted antibiotic-resistant pathogen, able to cause a broad spectrum of diseases in humans and animals17, 18. Based on the isolation origin, pathogenic variants are defined as intestinal pathogenic E. coli (InPEC) or extra-intestinal pathogenic E. coli (ExPEC). Furthermore, InPEC and ExPEC are subclassified into well-defined pathotypes according to disease manifestation, genetic background, phenotypic traits, and virulence factors (VFs)16,17,19.
Traditional culture and molecular techniques for pathogenic E. coli strains have allowed the rapid detection and identification of different pathotypes. However, they may be time-consuming, laborious, and frequently require high technical training19. Furthermore, no single method can be used to reliably study all pathogenic variants of E. coli because of the complexity of their genetic background. Currently, these drawbacks have been overcome with the advent of high-throughput sequencing (HTS) technologies. Whole-genome sequencing (WGS) approaches and bioinformatic tools have improved the exploration of microbial DNA affordably and at a large scale, facilitating the in-depth characterization of microbes in a single run, including closely related pathogenic variants20,21,22. Depending on the biological questions, several bioinformatics tools, algorithms, and databases can be used to perform data analysis. For instance, if the main goal is to assess the presence of ARGs, VFs, and plasmids, tools such as ResFinder, VirulenceFinder, and PlasmidFinder, along with their associated databases, might be a good starting point. Carriço et al.22 provided a detailed overview of the different bioinformatics software and related databases applied for microbial WGS analysis, from raw data preprocessing to phylogenetic inference.
Several studies have demonstrated the broad utility of WGS for genome interrogation regarding antimicrobial resistance attributes, pathogenic potential, and tracking of the emergence and evolutionary relationships of clinically relevant variants of E. coli sourced from diverse origins23,24,25,26. WGS has enabled the identification of molecular mechanisms underlying the phenotypic resistance to antimicrobials, including those rare or complex resistance mechanisms. This is through detecting acquired ARG variants, novel mutations in drug-target genes, or promoter regions27,28. Moreover, WGS offers the potential to infer antimicrobial resistance profiles without requiring prior knowledge about the resistance phenotype of a bacterial strain29. Alternatively, WGS has allowed the characterization of the mobile genetic elements (MGEs) carrying both antimicrobial resistance and virulence features, which has driven the bacterial genome evolution of existing pathogens. For instance, the application of WGS during the investigation of the German E. coli outbreak in 2011 resulted in uncovering the unique genomic features of an apparently novel E. coli pathotype; interestingly, those outbreak strains originated from the enteroaggregative E. coli (EAEC) group, which acquired the prophage encoding the Shiga toxin from the enterohemorrhagic E. coli (EHEC) pathotype30.
This work presents a methodological adaptation of the workflow for bacterial WGS using a benchtop sequencer. Moreover, a bioinformatics pipeline is provided using web-based tools to analyze the resulting sequences and further support researchers with limited or no bioinformatics expertise. The described methods allowed elucidation of the antimicrobial resistance, virulence, and mobilome features of a pathogenic E. coli strain ACM5, isolated in 2011 from inland farmed Oreochromis spp. in Sinaloa, Mexico12.
Protocol
NOTE: The E. coli strain ACM5 was recovered by processing and culturing the fish sample for fecal coliform (FC) determination12. During the fish sampling, fish did not show clinical signs of disease, bacterial, or fungal infection, and a mean temperature of 22.3 °C prevailed. After isolation, the E. coli isolate was subjected to biochemical testing and cryopreserved in brain heart infusion (BHI) broth with DMSO (8% v/v) as a cryoprotective agent.
1. Reactivation of frozen E. coli ACM5 stock culture
- From the frozen bacterial stock, open the tube and use a sterile loop, pipette tip, or toothpick to scrape the surface of the frozen bacterial culture.
- Streak the bacteria onto a Luria-Bertani (LB) agar plate and incubate at 37 ± 2 °C for 24 h.
2. Determination of antibacterial susceptibility
NOTE: The antimicrobial susceptibility testing described here corresponds to the disk diffusion method based on the Clinical and Laboratory Standards Institute (CLSI) guidelines (M02 Ed13:2018)31. E. coli ATCC 25922 strain is required for quality control purposes.
- Inoculum preparation by colony suspension method
- From the LB plate incubated in step 1.2, streak one or two colonies onto a Mueller-Hinton (MH) agar plate and incubate at 37 ± 2 °C for 18-24 h.
- Use a sterile loop to pick two or three colonies from the freshly sub-cultured E. coli isolate on MH agar plate and resuspend them in 3 mL of sterile MH broth or 0.85% (w/v) saline solution, mixing thoroughly by vortexing to obtain a uniform bacterial suspension.
- Using a UV-visible spectrophotometer (see Table of Materials), adjust the bacterial suspension to turbidity comparable to a 0.5 MacFarland standard (equivalent to approximately 1-2 x 108 cells/mL). If the bacterial suspension is too light or too heavy, add more E. coli colonies or MH broth as appropriate. Use the prepared inoculum within 15 min of preparation.
- Inoculation of the test agar plates
- Dip a sterile cotton swab into the adjusted bacterial suspension. Rotate the swab several times and press it firmly on the inside wall of the tube to remove excess fluid from the swab.
- Inoculate an MH agar plate by streaking the swab 3x over the entire agar surface, rotating the plate 60° each time. Swab the rim of the agar as well to ensure an even distribution of the inoculum.
- Leave the Petri dish lid ajar for 3-5 min to allow moisture to evaporate.
- Application of the antimicrobial disk onto inoculated agar plates
- Distribute evenly and press each antimicrobial disk (see Table of Materials) down onto the surface of the inoculated agar plates to ensure complete contact. Invert the plates within 15 min after disk application and incubate at 35 ± 2 °C for 16-18 h.
NOTE: A maximum of 12 and six disks must be used in Petri dishes of 150 mm and 100 mm, respectively.
- Distribute evenly and press each antimicrobial disk (see Table of Materials) down onto the surface of the inoculated agar plates to ensure complete contact. Invert the plates within 15 min after disk application and incubate at 35 ± 2 °C for 16-18 h.
- After incubation, measure the zone of inhibition sizes using a Vernier caliper and interpret the resulting diameters according to the CLSI breakpoint criteria (M100 - Table 2A)32.
3. Genomic DNA (gDNA) extraction and quantification
- Genomic DNA extraction
- Resuspend a loopful of freshly cultured E. coli colonies in 5 mL of LB broth and incubate overnight in a shaking incubator (180 rpm) at 37 ± 2 °C.
- Centrifuge the bacterial suspension at 3,500 x g for 5 min and carefully discard the supernatant.
- Extract gDNA following the DNA extraction kit guidelines (see Table of Materials).
- Check the purity of the gDNA by measuring optical density at 260/280 nm (ratio: >1.8) and 260/230 nm (ratio: 2.0-2.2) using a UV-visible spectrophotometer. Perform 0.8% agarose gel electrophoresis to verify the gDNA integrity.
- Genomic DNA quantification
NOTE: Use only thin-wall, clear, 0.5 mL PCR tubes approved by the fluorescence assay kit manufacturer (see Table of Materials).- Set up the required number of tubes for samples and assay standards. Prepare the working assay solution by mixing component A and component B provided in the kit, following the manufacturer's guidelines.
- Prepare the assay standards by adding 190 µL of the working solution and 10 µL of each standard to the appropriate tube (two standards are required).
- Add 198 µL of the working solution and 2 µL of the DNA sample to the appropriate tube. Vigorously mix by vortexing all the tubes for 5 s. Be careful not to create bubbles.
- Incubate all tubes for 2 min at room temperature, protected from light. Measure the fluorescence of all tubes based on the manufacturer's guidelines using the fluorometer (see Table of Materials).
- Properly adjust the gDNA sample concentration for sequencing proceeding.
4. DNA library preparation
NOTE: DNA library preparation and sequencing were performed following the manufacturer's guidelines and protocols (see Table of Materials). The starting gDNA concentration is 4.0 ng.
- Tagmentation, PCR amplification, and indexing
- In a 0.2 mL tube, add 2.5 µL of tagmentation buffer and 2 µL of input gDNA (2.0 ng/µL). Mix gently by pipetting.
- Add 1 µL of amplification buffer and mix gently by pipetting. Spin down at 280 x g for 1 min at room temperature.
- Place the samples in a thermocycler (see Table of Materials) and run the following PCR program: 55 °C for 5 min, then hold at 10 °C. When the samples reach 10 °C, proceed immediately to neutralize the reaction.
- Add 1 µL of neutralizing buffer to the sample tubes and mix gently by pipetting. Spin down at 280 x g for 1 min and incubate for 5 min at room temperature.
- Add 1.7 µL of each index adapter (i.e., indexes i7 and i5) and 3 µL of indexing PCR master mix. Mix gently by pipetting. Spin down at 280 x g for 1 min at room temperature.
- Place the samples in the thermocycler and perform a second PCR reaction as follows: 72 °C for 3 min, 95 °C for 30 s, 18 cycles of 95 °C for 10 s, 55 °C for 30 s, 72 °C for 30 s, 72 °C for 5 min, and hold at 10 °C.
- Amplified library clean up
- Mix by vortexing the commercial magnetic bead solution (see Table of Materials) based on the manufacturer's guidelines.
- Transfer the tagmented/indexed gDNA sample to a new 1.5 mL tube and add 0.6 µL of magnetic beads for each µL of the final volume of gDNA sample (≈13 µL). Mix gently by pipetting and incubate for 5 min at room temperature.
- Place the sample tubes on a magnetic rack (see Table of Materials) for 2 min until the supernatant has cleared. Remove and discard the supernatant carefully without disturbing the beads.
- Add 200 µL of freshly prepared 80% ethanol without mixing. Incubate for 30 s until the eluate clears and carefully remove and discard the supernatant without disturbing the beads.
- Perform a second wash step. Add 200 µL of 80% ethanol to the beads and incubate for 30 s. After the eluate clears, remove and discard the supernatant and air-dry the beads for 10 min.
- Add 15 µL of 10 mM tris buffer (pH 8) to the beads and mix gently by pipetting. Incubate for 2 min at room temperature. Place the sample tubes again on the magnetic rack for 2 min, allowing the supernatant to clear.
- Carefully transfer 14 µL of the supernatant from the sample tube to a new 0.2 mL tube. This is the cleaned-up library.
NOTE: At this point, the cleaned-up library can be stored at -20 °C for up to 7 days.
- Library normalization
- Proceed to quantify the cleaned-up libraries by fluorescence assay as described in step 3.2, except for step 3.2.5.
- Visualize the cleaned-up libraries on a 1% agarose gel electrophoresis to determine the average fragment sizes.
- Calculate the molarity value of the library using the following equation:
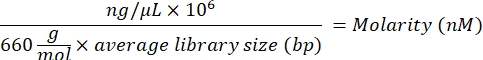
- Using the molarity value, calculate the proper volumes of resuspension buffer (RSB) and cleaned-up libraries to dilute the individual library at a starting concentration of 10 nM.
5. Library pooling, denaturalizing, and sequencer initiating
- Thaw the reagent cartridge following the manufacturer's directions. Extract the flow cell from the refrigerator (4 °C) and bring it to room temperature before sequencing.
- Pool 5 µL of each normalized library (10 nM) into a low-bind 1.5 mL tube. Dilute the pooled library at 4 nM with the proper volume of RSB.
- Add 5 µL of the pooled library (4 nM) and 5 µL of 0.2 N NaOH in a new low-bind 1.5 mL tube. Briefly mix by vortexing, spin down at 280 x g for 1 min, and incubate for 5 min at room temperature to denature the pooled library into single strands.
- Gently mix 10 µL of denatured pooled library and 990 µL of prechilled hybridization buffer by pipetting and place on ice until the final dilution for sequencer loading is performed. The concentration of the denatured pooled library is 20 pM.
- Thaw and prepare a control library (see Table of Materials) at a concentration of 4 nM by mixing 2 µL of control library (10 nM) and 3 µL of nuclease-free water.
- Mix 5 µL of the control library (4 nM) and 5 µL of freshly prepared 0.2 N NaOH. Briefly mix by vortexing, spin down at 280 x g for 1 min, and incubate for 5 min at room temperature to denature the control library into single strands.
- Gently mix 10 µL of denatured control library and 990 µL of prechilled hybridization buffer by pipetting and place on ice until the final dilution for sequencer loading is done. The concentration of the denatured control library is 20 pM.
- In a new low-bind 1.5 mL tube, combine 594 µL of the denatured pooled library at 20 pM and 6 µL of the denatured control library at 20 pM. Mix properly.
- Dilute the final library mix (20 pM) to a final loading concentration of 1.2 pM in a volume of 600 µL using the prechilled hybridization buffer.
- Before loading the final library mix onto the reagent cartridge, perform an additional thermal pretreatment to achieve efficient loading into the flow cell. Incubate the final library mix for 2 min at 96 °C. Invert the tube to mix, and place the tube on ice for 5 min.
- Load 500 µL of the final library mix into the designed reservoir on the reagent cartridge. Initiate sequencing following the guidelines. Load the flow cell and the reagent cartridge and set up the sequencing run.
6. Sequence data analysis
NOTE: Check Supplementary File 1 for further description of the general WGS data preprocessing, software, parameter settings, and sequence analysis of the E. coli genome.
- Connect to the sequencing data server and download the FASTQ files.
- Evaluate the initial quality of raw sequence data with third-party software. Remove remnant adapter sequences, low-quality bases (<Q30), and short reads (<50 bp) from the raw sequencing data (see Supplementary File 1).
- Assemble the quality-checked sequencing data into contig- or scaffold-level using third-party software (see Supplementary File 1).
- Perform genome annotation by submitting the FASTA file containing the assembled genome to the RAST server (https://rast.nmpdr.org/).
- Upload the FASTA file to the Center for Genome Epidemiology (CGE) (http://www.genomicepidemiology.org/services/) and ClermonTyping (http://clermontyping.iame-research.center/index.php) web platforms to identify epidemiological features, ARGs, VAGs, and plasmids (see Supplementary File 1).
- Upload the FASTA file to the PHASTER server to identify prophage sequences (https://phaster.ca/).
Results
The antimicrobial susceptibility was determined by the disk diffusion method and interpreted by CLSI breakpoint criteria for 12 antibiotics spanning six distinct antimicrobial classes, that is, aminoglycosides, β-lactams, fluoroquinolones, nitrofurans, phenicols, and folate pathway antagonists. The E. coli ACM5 exhibited sensitivity to all antibiotics except one β-lactam drug. Four β-lactam drugs were tested: ampicillin, carbenicillin, cephalothin, and cefotaxime. Among these, a 14 mm inhibition h...
Discussion
This study presents an adaptation of the bacterial WGS workflow using a benchtop sequencer and a pipeline for genomic characterization of a pathogenic E. coli variant. Depending on the sequencing platform used, the turnaround times (TATs) for wet laboratory procedures (bacterial culturing, gDNA extraction, library preparation, and sequencing) and sequence analysis could vary, particularly if slow-growing bacteria are studied. Following the protocol for WGS described above, the TAT was within 4 days, which is com...
Disclosures
The authors have nothing to disclose.
Acknowledgements
To the National Council of Science and Technology of Mexico (CONACyT by its acronym in Spanish) for the Doctoral scholarship awarded to José Antonio Magaña-Lizárraga [No. 481143].
Materials
| Name | Company | Catalog Number | Comments |
| Accublock Mini digital dry bath | Labnet | D0100 | Dry bath for incubation of tubes |
| Agencourt AMPure XP | Beckman Coulter | A63881 | Magnetic beads in solution for DNA library purification |
| DeNovix DS-11 | DeNovix Inc. | UV-Vis spectophotometer to check the quality of the gDNA extracted | |
| DNA LoBind Tubes | Eppendorf | 0030108418 | 1.5 mL PCR tubes for DNA library pooling |
| DynaMag-2 Magnet | Invitrogen, Thermo Fisher Scientific | 12321D | Magnetic microtube rack used during magnetic beads-based DNA purification |
| Gram-negative Multibac I.D. | Diagnostic reseach (Mexico) | PT-35 | Commercial standard antibiotic disks for antimicrobial susceptibility testing |
| MiniSeq Mid Output Kit (300-cycles) | Illumina | FC-420-1004 | Reagent cartdrige for paired-end sequencing (2x150) |
| MiniSeq System Instrument | Illumina | SY-420-1001 | Benchtop sequencer used for Next-generation sequencing |
| MiniSpin centrifuge | Eppendorf | 5452000816 | Standard centrifuge for tubes |
| Nextera XT DNA Library Preparation Kit | Illumina | FC-131-1024 | Reagents to perform DNA libraries for sequencing. Includes Box 1 and Box 2 reagents for 24 samples |
| Nextera XT Index Kit v2 | Illumina | FC-131-2001, FC-131-2002, FC-131-2003, FC-131-2004 | Index set A, B, C, D |
| PhiX Control v3 | Illumina | FC-110-3001 | DNA library control for sequencing |
| Precision waterbath | LabCare America | 51221081 | Water bath shaker used for bacterial culture |
| Qubit 1X dsDNA HS Assay Kit | Invitrogen, Thermo Fisher Scientific | Q33231 | Reagents for fluorescence-based DNA quantification assay |
| Qubit 2.0 Fluorometer | Invitrogen, Thermo Fisher Scientific | Q32866 | Fluorometer used for fluorescence assay |
| Qubit Assay tubes | Invitrogen, Thermo Fisher Scientific | Q32856 | 0.5 mL PCR tubes for fluorescence-based DNA quantification assay |
| SimpliAmp Thermal Cycler | Applied Biosystems, Thermo Fisher Scientific | A24811 | Thermocycler used for DNA library amplification |
| Spectronic GENESYS 10 Vis | Thermo | 335900 | Spectophotometer used for bacterial suspension in antimicrobial susceptibility testing |
| ZymoBIOMICS DNA Miniprep Kit | Zymo Research Inc. | D4300 | Kit for genomic DNA extraction (50 preps) |
References
- Naylor, R. L., et al. A 20-year retrospective review of global aquaculture. Nature. 591 (7851), 551-563 (2021).
- Quesada, S. P., Paschoal, J. A. R., Reyes, F. G. R. Considerations on the aquaculture development and on the use of veterinary drugs: special issue for fluoroquinolones-a review. Journal of Food Science. 78 (9), 1321-1333 (2013).
- Defoirdt, T., Sorgeloos, P., Bossier, P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Current Opinion in Microbiology. 14 (3), 251-258 (2011).
- Stentiford, G. D., et al. New paradigms to help solve the global aquaculture disease crisis. PLOS Pathogens. 13 (2), 1006160 (2017).
- Chen, H., et al. Tissue distribution, bioaccumulation characteristics and health risk of antibiotics in cultured fish from a typical aquaculture area. Journal of Hazardous Materials. 343, 140-148 (2018).
- Zhou, M., et al. Antibiotics control in aquaculture requires more than antibiotic-free feeds: A Tilapia farming case. Environmental Pollution. 268, 115854 (2021).
- Feng, Y., et al. Ecological effects of antibiotics on aquaculture ecosystems based on microbial community in sediments. Ocean & Coastal Management. 224, 106173 (2022).
- Shen, X., Jin, G., Zhao, Y., Shao, X. Prevalence and distribution analysis of antibiotic resistance genes in a large-scale aquaculture environment. Science of The Total Environment. 711, 134626 (2020).
- Su, H., et al. Contamination of antibiotic resistance genes (ARGs) in a typical marine aquaculture farm: source tracking of ARGs in reared aquatic organisms. Journal of Environmental Science and Health, Part B. 55 (3), 220-229 (2020).
- Oliveira, R. V., Oliveira, M. C., Pelli, A. Disease infection by Enterobacteriaceae family in fishes: a review. Journal of Microbiology & Experimentation. 4 (5), 00128 (2017).
- Barbosa, M. M. C., et al. Sorologia e suscetibilidade antimicrobiana em isolados de Escherichia coli de pesque-pagues. Arquivos do Instituto Biológico. 81 (1), 43-48 (2014).
- Valenzuela-Armenta, J. A., et al. Microbiological analysis of Tilapia and water in aquaculture farms from Sinaloa. Biotecnia. 20 (1), 20-26 (2018).
- Reza, R. H., Shipa, S. A., Naser, M. N., Miah, M. F. Surveillance of Escherichia coli in a fish farm of Sylhet, Bangladesh. Bangladesh Journal of Zoology. 48 (2), 335-346 (2021).
- Liao, C. -. Y., et al. Antimicrobial resistance of Escherichia coli From aquaculture farms and their environment in Zhanjiang, China. Frontiers in Veterinary Science. 8, 806653 (2021).
- Dewi, R. R., et al. Prevalence and antimicrobial resistance of Escherichia coli, Salmonella and Vibrio derived from farm-raised Red Hybrid Tilapia (Oreochromis spp.) and Asian Sea Bass (Lates calcarifer, Bloch 1970) on the west coast of Peninsular Malaysia. Antibiotics. 11 (2), 136 (2022).
- Leimbach, A., Hacker, J., Dobrindt, U. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Current Topics in Microbiology and Immunology. 358, 3-32 (2013).
- Kaper, J. B., Nataro, J. P., Mobley, H. L. T. Pathogenic Escherichia coli. Nature Reviews Microbiology. 2 (2), 123-140 (2004).
- Croxen, M. A., Finlay, B. B. Molecular mechanisms of Escherichia coli pathogenicity. Nature Reviews Microbiology. 8 (1), 26-38 (2010).
- Croxen, M. A., et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clinical Microbiology Reviews. 26 (4), 822-880 (2013).
- Bertelli, C., Greub, G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clinical Microbiology and Infection. 19 (9), 803-813 (2013).
- Lynch, T., Petkau, A., Knox, N., Graham, M., Van Domselaar, G. A primer on infectious disease bacterial genomics. Clinical Microbiology Reviews. 29 (4), 881-913 (2016).
- Carriço, J. A., Rossi, M., Moran-Gilad, J., Van Domselaar, G., Ramirez, M. A primer on microbial bioinformatics for nonbioinformaticians. Clinical Microbiology and Infection. 24 (4), 342-349 (2018).
- Magaña-Lizárraga, J. A., et al. Draft genome sequence of Escherichia coli M51-3: a multidrug-resistant strain assigned as ST131-H30 recovered from infant diarrheal infection in Mexico. Journal of Global Antimicrobial Resistance. 19, 311-312 (2019).
- Pérez-Vázquez, M., et al. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. Journal of Antimicrobial Chemotherapy. 74 (12), 3489-3496 (2019).
- Massella, E., et al. Snapshot study of whole genome sequences of Escherichia coli from healthy companion animals, livestock, wildlife, humans and food in Italy. Antibiotics. 9 (11), 782 (2020).
- Magaña-Lizárraga, J. A., et al. Genomic profiling of antibiotic-resistant Escherichia coli isolates from surface water of agricultural drainage in north-western Mexico: detection of the international high-risk lineages ST410 and ST617. Microorganisms. 10 (3), 662 (2022).
- Saracino, I. M., et al. Next Generation sequencing for the prediction of the antibiotic resistance in Helicobacter pylori: a literature review. Antibiotics. 10 (4), 437 (2021).
- Ghosh, A., Saha, S. Survey of drug resistance associated gene mutations in Mycobacterium tuberculosis, ESKAPE and other bacterial species. Scientific Reports. 10 (1), 8957 (2020).
- Su, M., Satola, S. W., Read, T. D. Genome-based prediction of bacterial antibiotic resistance. Journal of Clinical Microbiology. 57 (3), 01405-01418 (2019).
- Brzuszkiewicz, E., et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC). Archives of Microbiology. 193 (12), 883-891 (2011).
- . CLSI Performance Standards for Antimicrobial Disk Susceptibility Tests. 13th ed. CLSI standard M02. Wayne, PA: Clinical and Laboratory Standards Institute Available from: https://clsi.org/standards/products/microbiology/documents/m02/ (2018)
- CLSI Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100. Clinical and Laboratory Standards Institute Available from: https://clsi.org/standards/products/microbiology/documents/m100/ (2021)
- Ewing, B., Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 8 (3), 186-194 (1998).
- Quainoo, S., et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clinical Microbiology Reviews. 30 (4), 1015-1063 (2017).
- Desai, A., et al. Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS ONE. 8 (4), 60204 (2013).
- Nishino, K., Yamada, J., Hirakawa, H., Hirata, T., Yamaguchi, A. Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to β-lactams. Antimicrobial Agents and Chemotherapy. 47 (9), 3030-3033 (2003).
- Li, M., et al. The resistance mechanism of Escherichia coli induced by ampicillin in laboratory. Infection and Drug Resistance. 12, 2853-2863 (2019).
- Ménard, L. -. P., Dubreuil, J. D. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Critical Reviews in Microbiology. 28 (1), 43-60 (2002).
- Dubreuil, J. D. EAST1 toxin: An enigmatic molecule associated with sporadic episodes of diarrhea in humans and animals. Journal of Microbiology. 57 (7), 541-549 (2019).
- Goldstein, S., Beka, L., Graf, J., Klassen, J. L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genomics. 20 (1), 23 (2019).
- Guerrero, A., Gomez-Gil, B., Lizarraga-Partida, M. L. Genomic stability among O3:K6 V. parahaemolyticus pandemic strains isolated between 1996 to 2012 in American countries. BMC Genomic Data. 22 (1), 38 (2021).
- FAO Applications of Whole Genome Sequencing (WGS) in food safety management. Food and Agriculture Organization of the United Nations Available from: https://www.fao.org/documents/card/es/c/61e44b34-b328-4239-b59c-a9e926e327b4/ (2016)
- Rantsiou, K., et al. Next generation microbiological risk assessment: opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. International Journal of Food Microbiology. 287, 3-9 (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved