Method Article
利用3D打印技术来合并MRI与组织学:一个大脑切片协议
摘要
该协议的总体目标是通过创建定制3D印刷脑持有者和切片机盒的精确对准与组织切片的磁共振成像(MRI)图像的卷。
摘要
Magnetic resonance imaging (MRI) allows for the delineation between normal and abnormal tissue on a macroscopic scale, sampling an entire tissue volume three-dimensionally. While MRI is an extremely sensitive tool for detecting tissue abnormalities, association of signal changes with an underlying pathological process is usually not straightforward. In the central nervous system, for example, inflammation, demyelination, axonal damage, gliosis, and neuronal death may all induce similar findings on MRI. As such, interpretation of MRI scans depends on the context, and radiological-histopathological correlation is therefore of the utmost importance. Unfortunately, traditional pathological sectioning of brain tissue is often imprecise and inconsistent, thus complicating the comparison between histology sections and MRI. This article presents novel methodology for accurately sectioning primate brain tissues and thus allowing precise matching between histology and MRI. The detailed protocol described in this article will assist investigators in applying this method, which relies on the creation of 3D printed brain slicers. Slightly modified, it can be easily implemented for brains of other species, including humans.
引言
In vivo MRI provides a noninvasive and sensitive measure of tissue integrity at the macroscopic level. Changes in MRI signal intensity seen in vivo are outcome measures in many ongoing clinical trials.1 While the intensity changes seen via MRI can identify areas of abnormality in the context of the whole brain, they are often not sufficiently specific to differentiate pathological processes. This is especially true of dynamic processes involving multiple pathologies. For example, in multiple sclerosis (MS) or its animal model, experimental autoimmune encephalomyelitis (EAE), inflammation, edema, myelin degradation, axonal destruction, gliosis, and neuronal death overlap. 2, 3 To obtain the necessary specificity regarding the underlying pathology, context must be taken into account, together with knowledge of the histology of the MRI-identified abnormal tissues.
However, even in well-controlled animal experiments, matching histology with in vivo MRI is fundamentally challenging for various reasons. First, the difference in dimensional scales between histology sections and MRI is of several orders of magnitude.4 Second, for proper comparison, the orientation of MRI slice plane must match the sectioning plane of the brain tissue when cut. Due to the shape of the brain, it is very difficult to make consistently straight and accurate cuts when the brain is sitting on a flat surface. Third, the large size of the brain relative to a potentially small area of interest (lesion, tumor, etc.) creates a "needle-in-a-haystack" scenario for the pathologist processing the tissue. Fourth, even when the target tissue is found, it is commonly processed in such a way as to render virtually impossible an association with the original MRI data. Finally, traditional pathological sectioning of brain tissue is often imprecise and inconsistent, further complicating the comparison between histology sections and MRI images.
Previous attempts to overcome these challenges relied on the use of deformational algorithms to coregister the data and/or placement of fiducial markers within or around the tissue as a reference.5, 6, 7, 8 The former approach requires complex computational models that are particularly susceptible to complications due to data formatting, imaging artifacts, and changes caused by tissue processing.4 On the other hand, the latter approach introduces the possibility of contaminating or otherwise harming the tissue itself.9
The approach described here improves the transition between modalities through the use of postmortem MRI to bridge the gap between in vivo MRI and histology. Postmortem MRI provides three-dimensional (3D) images of the brain at higher resolution than can be achieved in vivo and furthermore provides the data needed for producing a morphologically accurate model of the brain surface. This digital model can then be used to create a 3D-printed custom holder for the brain. With careful positioning, the brain holder allows for precise, MRI-oriented brain sectioning, reducing the need for complex mathematical algorithms, and enables a focus on specific regions for targeted sampling.
Our laboratory recently introduced new methods for creating custom brain holders and slicers using postmortem MRI and 3D-printing technology for human10 and marmoset brains.4 The two methods allow for a more accurate correlation between MRI and histology in a research setting, and ultimately allow a deeper understanding of the specific pathology underlying MRI abnormalities. Carefully designed experiments, in which the brain is sampled repeatedly over time in vivo, can provide context for interpretation of the pathology, which in turn can add specificity to interpretation of the MRI. Here, we present a modified protocol in a unified framework that can be applied to any brain tissue, whether it derive from nonhuman primates, rodents, or humans. We provide detailed instructions, and a corresponding video, for the marmoset sectioning. Although the overall protocol applies to any type of brain, due to differences in MRI acquisition and tissue size, as well as the challenges encountered when dealing with specific brain types, there are some differences in the approach depending on the type of brain being processed. In this presentation, sections with "human" will denote differences in protocol specific to the human brain.
研究方案
所有动物处理和本文中所描述的程序是按照由神经疾病的国家研究所和中风动物护理和使用委员会批准的方案进行。脑从诱导培养EAE普通狨猴( 普通狨猴 )收集。 11脑储存在10%福尔马林3周和4%多聚甲醛穿心灌注安乐死后一年之间。
1.死后MRI准备与收购
- 狨猴脑
- 准备用棉花纱布一个工作站,在50ml离心管中,小铲,约30毫升氟化油,石蜡,和狨猴脑。
- 装满氟化油纱布20毫升标记管。压缩纱布沿途去除气泡。
- 从用纸巾大脑的表面轻轻地干燥福尔马林。插入脑内朝向底部的额极管。小心使用双方围绕多个纱布固定其位置固定的大脑在管。参阅补充部分11用于创建的额外MRI扫描设置的MRI脑支架的方法。
- 填用纱布和氟化油的管子的其余部分。小心地除去气泡一路走来。固定帽和用石蜡密封该管。
- 马克与纵裂的线帽。包裹管中纸巾并将其插入带有标志顶部中心的线圈。
- 获取2D自旋回波T2。参数在表1中给出。
- 打开10解剖150微米T2-weigted收购Mipav并注册到6 日的收购。
注:登记是一个优化的自动登记3.5D算法与9个自由度,窗正弦内插,归一化互相关的成本函数,具有一个鲍威尔的主叫Brent的搜索算法。轮换从+10取样6;至-10℃粗旋转递增5度和精细旋转递增1°。然后平均注册的图像:公用事业,图像计算器,批量图片,平均。
- 人脑
- 从利用在中脑的水平切割脑干分离前脑。 10半球也可以用一个切下来中线分开。
- 在圆筒形管中的前脑在一端半球形穹顶和在另一端的喷口的位置。
- 通过喷口填充用氟化油的管。通过使用喷温和的吸力约30分钟去除气泡。
- 获得3D T1-MPRAGE。参数在表1中给出。
2.提取脑表面:Mipav 7.2
- 打开冠状方向的MRI检查。
- 选择算法,转换工具,转换,重新取样。选择用户自定义大小和重采样ISOTropic像素:0.1×0.1×0.1毫米。保存重采样MRI作为Brain_MRI_Resampled。人类:要重新取样各向同性体素0.3×0.3×0.3毫米。
- 选择算法,过滤器(空间),非线性降噪。使用默认设置,单击确定。
- 选择显示查找表,然后点击双阈值的按钮。拖动图上的滑块来覆盖整个大脑。
- 选择算法,分割,阈值,使用阈值最小值/最大值。进入位于强度图(只是规模以下)进入"下限"框左下角的价值。选择在输出图像类型"二进制",取消选择"反转门槛"。
- 选择算法,形态,补洞。检查"过程中的2.5D"。
- 因为这是满脑的MRI,存在需要填充后脑和皮质之间的一些空的空间。人类的大脑:跳过此步骤。
- 使用线VOI,绘制一个连接在最外侧点后脑和皮质的脑的两侧之间。通过大脑继续此。
- 选择VOI,VOI转换,全部以二进制掩码。选择实用程序,图像计算器。选择或从操作下拉菜单并选择大脑面具。选择"促进目标图像类型"
- 选择算法,形态,补洞。选中"在2.5D过程"
- 保存二进制掩码为Brain_Model.nii。
- 选择算法,提取表面(移动立方体)。选择面膜的形象,另存为Brain_Model.ply。
3.选择切片的位置:Mipav 7.2
- 确定的利益或起始位置组织。计算预期的楼板厚度。在狨猴脑,计算每节30 MRI片,每刀片间隙5 MRI片,创建一个具有0.5毫米刀片间隙3毫米段,导致〜3.5毫米板。人类的大脑:每部20 MRI片和4 MRI SL每个刀片间隙冰,创建了1.2毫米刀片间隙6毫米部分,导致〜7.2毫米板。
- 在第一刀片间隙的位置,点击"盒子VOI",然后画一个框在大脑。点击框内的选择它。复制和粘贴对应于叶片间隙的每个的MRI切片的轮廓。
- 由对应于部分厚度的MRI切片的数目跳到复制并粘贴对应于下一个叶片间隙轮廓。通过大脑重复此过程。
- 选择VOI,VOI转换,所有的二进制掩码。另存为Blade_Gaps.nii。
- 选择算法,抽离曲面(移动立方体),面膜的形象,Blade_Gaps.ply。
4.创建MRI刀片地图:Mipav 7.2
- 打开Brain_MRI_Resampled和Blade_Gaps.nii图像。
- 在选中Blade_Gaps.nii图像,请选择实用程序,图像数学。选择Promote图像类型和繁殖。输入10000作为值。
- 选择实用程序,图像计算器。选择添加,然后选择从图像下拉框中Brain_MRI_Resampled形象。选择促进目标图像类型。
- 将图像保存为Brain_BladeMap.nii。
- 通过点击三平面视图,其中该大脑将切片的位置可以在三个正交视图中看到。
5.导入大脑和刀片间隙表面:Netfabb专业
- 选择部分,添加一部分。选择文件Brain_Model.ply和Blade_Gaps.ply
- 选择大脑和点击修复模式。在修复模式:
- 点击外壳选择按钮,然后点击大脑。
- 点击切换选择键选择其他网格。点击删除,以删除其他网格。
- 点击应用修复,并删除旧的一部分。
- 右键单击大脑。选择移动。点击原点按钮。记录显示的XYZ参数。这些翻译参数是需要保持刀片定位在Mipav成立。点击翻译。然后关闭窗口。 (不要点击翻译不止一次,这将转化再次使用相同的参数。)
- 选择Blade_Gaps模式。右键单击零件,然后选择移动。输入在XYZ参数框先前记录的XYZ值。现在点击翻译并关闭窗口。
- 选择Brain_Model模型,点击修复模式。在修复模式:
- 点击外壳选择按钮,然后点击大脑。
- 右键单击并选择平滑三角形。进入4-5迭代。检查防止成交量萎缩。人类的大脑:1-2迭代。
- 点击右键,选择缩小的三角形。输入目标三角形数量200000单击执行。
- 点击自动修复,默认的修复。然后点击应用修复
- 右击,重命名。重命名平滑大脑Smoothed_Brain_Model。
- 选择Smoothed_Brain_Model。右键单击,导出,STL。
6.编辑脑轮廓:Meshmixer
- 导入Smoothed_Brain_Model到Meshmixer。
- 用雕刻和选择工具来调整网格。编辑包括:
- 使用雕刻工具,乐百氏顺利。平滑对应于该线的VOI的区域。人类的大脑:跳过此步骤。
- 平滑,将在框朝下皮层的表面上。
- 抚平可能已在网格划分和编辑过程中创建的小凹坑。
- 选择分析,检查,Autorepair所有。
- 导出Smoothed_Brain_Model为Smoothed_Edited_Brain_Model。
7.创建脑切片盒:Netfabb专业
- 选择部分,添加部分。选择FILËSmoothed_Edited_Brain_Model。
- 选择部分,添加部分。然后选择STL文件切片脑和Parts_Marmoset点击打开。人类的大脑: 脑切片Parts_Human(补充代码文件)。
- 右键单击零件,然后选择扩展,炮弹零件。分别选择各个部分,然后点击鼠标右键重命名。 (点击旁边的一个对象眼睛隐藏它,或使其可见)。 重命名大箱主 ,小盒子Sub和切割盒Box_Cutout,六角形状Blade_Holder_Main,小扁盒刀片切片和半管对象摇篮 。人类的大脑:没有Blade_Holder_Main, 切片机刀片 ,或摇篮 。
- 隐藏主 , 次 , 刀片,Blade_Holder_Main, 切片机刀片 ,和摇篮和使用按住Shift键选择来选择所有六个和Box_Cutout。只有Box_Cutout和Smoothed_Edited_Brain_Model应该是可见的,但Smoothed_Edited_Brain_Model不应该被选中。
- 单击并拖动选定的部分调整相对于大脑中的盒位置。
- 大脑在盒子的中心位置。当前位置在框中深足的大脑被紧紧揪住,但不宜过深,以创建一个防止妥善安置出挑。
- 一旦定位,大脑盒轮廓可以为突出端进行测试。选择Box_Cutout和Smoothed_Edited_Brain_Model。选择布尔运算。
- 点击Smoothed_Edited_Brain_Model把它红色。
- 选择布尔减,并应用计算。
- 检查脑框将防止脑组织被安全地放入盒子突出端。如果这些出挑都存在,调节大脑所以它是方框内较浅。如果大脑在所需的深度和悬都存在,请参阅补充第10条的解决方案去除出挑。
- 选择Blade_Gaps模式。点击右键,选择移动。记录位置的Z值,然后关闭窗口。这将是最后叶片间隙的位置。
- 选择从大脑切片而来, 刀片 STL。点击右键,选择移动。
- 选择绝对平移。输入从7.8 Z值。对于X和Y值,输入从当前位置参数框对应的值
- 选择刀片 STL。点击右键,点击复制。
- 检查安排部分。在总计数进入叶片的总数量。输入相同的号码到Z计数盒。输入板坯厚度为Z轴的差距。点击复制。如果板坯厚度是变化的,叶片将需要单独地定位。从以前的每一个重复新刀片(从后移动到前)与z差距等于截面厚度该节。
- 通过重复步骤7.9和7.10的切片机刀片在完全相同的时间间隔如在切片机刀片的切片机刀片的部分的位置。人类的大脑:跳过此步骤。
- 选择切片机刀片与Blade_Holder_Main部分沿,并选择布尔运算。
- 在选择刀片切片所有刀片以突出他们的红色。点击布尔减,并应用计算。
- 选择修复模式。执行扩展的修复。选择适用的修复,并删除旧的一部分。
- 重命名部分刀架 。导出一部分STL。
- 按住Shift键选择Smoothed_Edited_Brain_Model,都在一步中创建的叶片模型,和小组和主 。点击布尔运算。
- 让所有部件除非本身主要红色lecting并且点击下的绿色框上的箭头将其移动到红色。
- 选择布尔减,然后选择适用的计算。
- 点击修复模式。在修复模式:
- 检查悬和尖点的框。这些可以在Netfabb或Meshmixer平滑。
- 单击自动修复,修复扩展。然后应用修复以及删除旧的一部分
- 右键单击修复大脑盒子并重新命名为脑切片机盒 。导出为STL。
8.打印脑切片机盒上的Ultimaker 2
- 绒猴脑:库拉索
- 导入大脑切片盒到库拉索。
- 选择旋转并拖动圆圈所以它平放在床上旋转框。
- 调整打印设置:0.1毫米层的分辨率,50%的填充密度,筏。
- 选择保存刀具轨迹SD卡。 (打印时间〜12小时)。
- 进口刀架在到库拉索并旋转它,因此槽上侧面和六边形面是在XZ或YZ平面。
- 复制的对象。
- 调整打印设置为0.2mm层的分辨率,20%的填充密度,满满当当。
- 选择保存刀具轨迹SD卡。 (打印时间〜3小时)
- 人脑:库拉索
- 导入大脑切片盒到库拉索和旋转8.1.2。
- 调整打印设置:0.2毫米层的分辨率,30-35%的填充密度,筏。
- 选择保存刀具路径到SD卡。 (打印时间为单箱半球〜70小时。)
- 在Ultimaker 2
- 应用胶水粘的胶水一层薄薄建造板。
- 插入SD卡。选择打印,然后选择一部分。
9.切脑
- 狨猴脑
- 制备工作站与固定大脑,大脑切片机,两个叶片座,切片机刀片,将1ml氟化油中,f纬度镊子,防护手套,和嵌入录像带。
- 将新的刀片切片机将在刀架上的插槽。确保每个刀片的斜边朝向在相同的方向。 处理切片机刀片时戴上防护手套。
- 从福尔马林中取出脑组织,并轻轻擦干。
- 将脑入切片机。氟化油几滴可以应用于脑和限幅器,以允许容易的定位。确保大脑牢固。
- 刀片持有人在相应的刀片插槽刀片位置。
- 向下推刀架坚决和应用缓慢的平衡压力通过大脑进行切割。
- 一次删除每个切片,一,从大脑的前部开始。它有助于除去板坯本身之前去除板坯前方切片机刀片。狠抓各板的前/后方向。
- 就拿前的照片每个板d的后表面。后板将最有可能包含独立的部分,因此要注意碎片的方向嵌入。将每个板成包埋盒,并把他们都变成10%的福尔马林溶液。
- 人脑
- 仔细测试大脑的配合在框中。
- 切片大脑从一端使用有角度的切割开始,缓慢而坚定地切片。通过每个刀片间隙切开大脑。
- 一次删除各平板之一,密切关注数量和每个板的前/后方向。
- 取各板坯的前,后表面的照片。放置在密封的10%福尔马林袋板坯。组织块将从板坯进行切割,并放置在盒嵌入。
10.大脑箱中取出出挑(补充部分)
- 拔出片:Meshmixer
- 导入Smoothed_Edited_Brain_Model。
- 选择编辑,制作切片。在制作切片:
- 选择3D堆叠,Z,进入1-2毫米的厚度。点击计算。当片加载,单击接受。
- 选择1或2片与邻近的皮质底部大周长。这些片应低于小组方块的水平。
- 导出每个切片作为Brain_Slice_#的。
- 扩展切片,将其取出出挑:Netfabb专业
- 导入Brain_Slice_#片。
- 每个重复#Brain_Slice_(如果选中取消选中安排部分)。
- 右键单击每个Brain_Slice_#,并选择分段的一个副本。
- 取消选中固定的缩放比率。然后扩大脑切片在Y方向上,这样它会到达子盒的底部的水平。
- 重命名这些片Brain_Slice_Big_#。
- 检查Y位置Ø˚F通过在部分右击并选择移动原始Brain_Slice_#。记录每个原始Brain_Slice_#片的Y位置。
- 进行计算:Brain_Slice_#[Y位置] - (Brain_Slice_Big_#[Y尺寸] - Brain_Slice _#[Y尺寸])
- 分别选择每个Brain_Slice_Big_#中,右键单击并选择移动。
- 从输入10.2.6中的Y平移参数框计算出的值。为X和Z平移参数,进入位于当前位置参数框中的值。选择绝对平移。点击翻译,然后关闭窗口。
注:Brain_Slice_Big_#切片将与脑和刀片沿使得当盒被减去。
- 从输入10.2.6中的Y平移参数框计算出的值。为X和Z平移参数,进入位于当前位置参数框中的值。选择绝对平移。点击翻译,然后关闭窗口。
11.狨猴脑MRI摇篮附加扫描
- 创建脑MRI摇篮
- 确保在Cr的顶部表面adle是在相同的高度的Box_Cutout。在摇篮的深度和大脑的位置应设置只是因为它是为限幅器。
- 按住Shift键选择来选择Smoothed_Edited_Brain_Model和摇篮 。
- 选择布尔运算。选择大脑突出显示红色,然后选择布尔减。然后应用计算。 (也选择(如果适用)Brain_Slice_Big_#切片。)
- 进入修复模式如先前的切片机完成在摇篮轮廓以除去任何尖锐点。选择扩展修复。应用修复,删除旧的一部分。
- 右击要重命名的部分脑部MRI摇篮 。选择导出,STL。
- 打印摇篮:库拉索
- 导入的MRI脑摇篮到库拉索并且使得与大脑切口平坦部朝上旋转。
- 调整打印设置:0.1毫米层的分辨率,100%的填充密度,筏。
- 选择保存刀具路径到SD卡。 (打印时间〜10小时)
- 在8.3中描述的Ultimaker 2打印。
- 采用从摇篮获取高分辨率T2 * MRI
- 从用纸巾大脑的表面轻轻地干燥福尔马林。
- 作为切片描述摇篮大脑的位置。
- 滑动脑和摇篮到50ml离心管中。它填充用氟化油的边缘。
- 轻轻挤压管,以允许气泡从大脑逸出。将帽插入到管帽的插图,以防止形成气泡那里。固定帽,并用石蜡密封该管。
- 如前所述放置管插入线圈。 3D T2 *参数在表1中给出。
- 打开18解剖100微米T2 *加权收购Mipav并注册到10 日的收购。登记参数是相同1.1.7。平均注册的图像:UTilities,图像计算器,批量图片,平均。
结果
该方法的流程被总结于图1。一旦脑切片,板坯的浅表面的MR图像和图片之间的视觉比较显示在多个板坯的好取向匹配(图2)。的板坯石蜡包埋后,它们被剖开上的切片和染色。高分辨率死后MRI和染色组织学切片之间的更彻底的比较表明横跨狨大脑的所有结构的精确和一致的匹配(图3)。
在MS的动物模型,动物发展脑白质病变整个脑白质传播。这些病变可通过无创性MRI表现进行检测。图4表明该技术的阐明MRI表现的病理基板的能力。 对体内 MRI检测微小病灶可以在两个死后MRI和组织学进行跟踪。如插图所示,病灶内脱髓鞘是驱动该MR信号变化(高信号相比周围组织)的主要成分之一。组织学和死后的MRI还可以显示病变错过对体内 MRI(图4)。
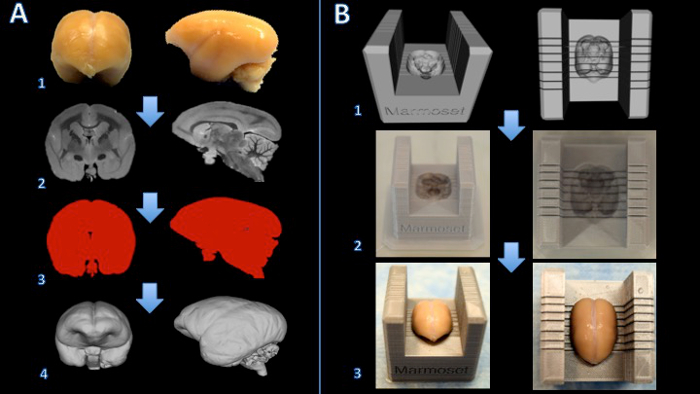
图1的工作流程创建狨脑切片机框。脑被固定用福尔马林(A1)和一T2加权的MRI用的每边为150μm(A2)中各向同性体素获得。图像处理和阈值处理,以创建一个二进制掩码(A3)。该表面然后在3D建模软件(A4)呈现。切片机模板和大脑模型之间的布尔减造成大脑切片机(B1)的数字模型。大脑切片机盒印刷三维打印机(B2)上。大脑然后用力放置在切片机框切削(B3)。 请点击此处查看该图的放大版本。
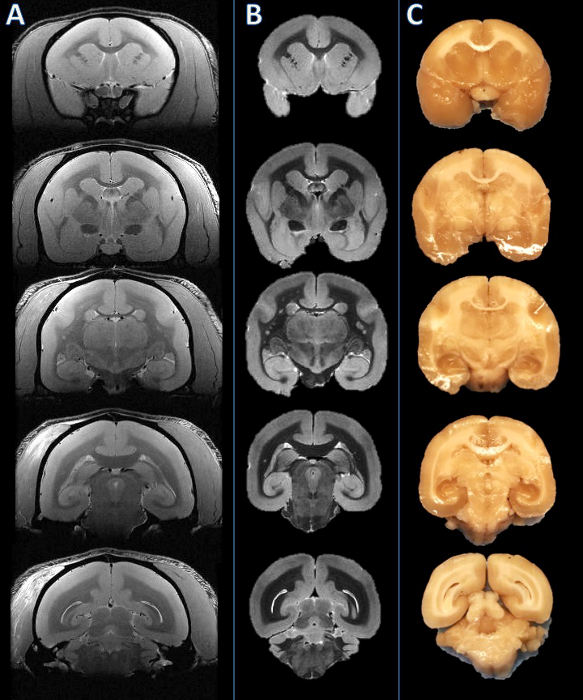
图2.从左至右:体内 MRI,MRI尸检和组织板的照片。相比于相应体内 MRI切片(A)的基础上死后的MRI(B)的切片平面建立和视觉。然后将脑切,并且发现所得板坯要一致(C)。 请点击此处查看该图的放大版本。
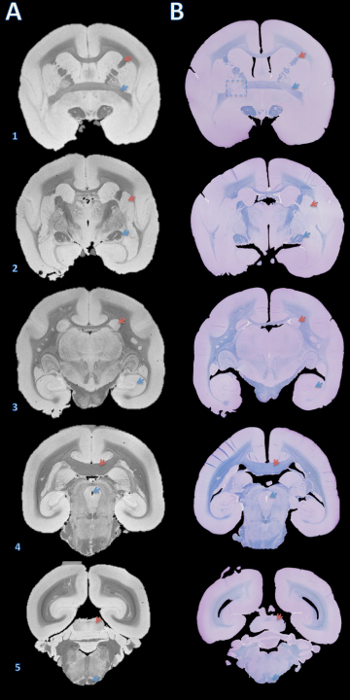
图3.高分辨率死后MRI和组织学部分匹配。板坯包埋在石蜡中,切成用切片机成4微米的部分,并具有快速蓝和甲酚紫(B)中染色。切片,然后在视觉上与100微米T2 *加权MRI基于大脑结构(A)相匹配。获取此图像细节在协议中和表1大脑结构的补充部分:(1)红色箭头=内囊,蓝色箭头=前联合; (2)红色箭头=壳,蓝色箭头=视束; (3)红色箭头=尾,蓝色箭头=海马; (4)红色箭头=胼胝体,蓝色箭头=脑导水管; (5)红色箭头=下丘,蓝色箭头=锥体束。在B 1中的虚线框表示,其中,无论是在脑切割或石蜡包埋,一个错误引起的绕Y轴的轻微旋转,从而导致在左侧的前联合的失配一个切片。 请点击这里查看更大的版本这个数字。
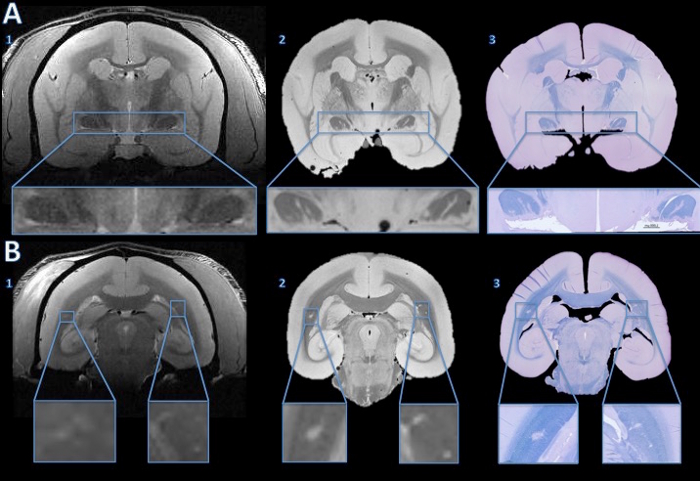
图4.跟踪从体内 MRI病变组织学切片。 体内 MRI显示异常高信号信号没有令人信服的证据表明,在任一视束(A1)病变。然而,高分辨率死后MRI显示两个视束(A2)超清晰的线条强烈。一个4微米的组织学切片的快速蓝/甲酚紫染色显示上看到体外的MRI高信号区脱髓鞘(A3)。在脑白质, 体内 MRI显示出微妙的双边高信号(B1,放大的插图)。该高信号区域的高分辨率死后MRI(B2)更为明显。一个4μm的组织学部分的LFB染色表明,这些区域被脱髓鞘(B3)。与基线相比我后Ñ体内MRI和一个苏木和-曙红染色,右侧被确定为解剖学异常,而不是一个脱髓鞘病灶。 请点击此处查看该图的放大版本。

补充代码文件。 Brain_Slicer_Parts_Marmoset.stl: 请点击这里下载该文件。 Brain_Slicer_Parts_Human.stl: 请点击这里下载该文件。 Cap_Insert.stl: 请点击这里下载该文件。
讨论
这里列出的协议使MRI和组织切片之间准确的比较。的协议,提出在可应用于人类或小动物,例如狨猴或啮齿类动物的大脑的统一格式。具体到大(人)和小(非人灵长类和啮齿类动物)的大脑突出显示,并在所附的视频和附图的差异,我们证明在狨猴中的应用。虽然该方法是简单的,该方法需要许多步骤以及使用多种类型的软件。此外,一些问题可能会影响这种方法的准确度是很重要的就更不用说了。
体内 MRI的图像质量是一个重要因素。为了最大限度地减少在MRI和数字化的组织学图像之间的图像分辨率的视差,应该使用尽可能小的磁共振成像的体素尺寸。这个概念也适用于死后的MRI的图像质量。虽然增加在死后的MRI采集时间允许更高的图像分辨率,该制剂可以引入的图像伪影,例如有关气泡焦信号丢失。这些文物可以掩盖该组织区域以及影响其轮廓。此外,在尸检的MRI的组织的尺寸很可能是由在固定过程和持续时间的影响。而在体内 体外 MRI匹配可以通过采集期间利用在切片几何设置解剖标志紧密近似非线性登记将仍然是必要的,相匹配这两个MRI图像达到更高的精确度。
大脑保持器和限幅器的设计也是一个关键步骤。在创建脑的数字模型,平滑算法应用于该相对于固定脑模型稍微放大。这使大脑容易插入到其支架和限幅器,并降低在支架锐利边缘及#39; S轮廓。但是,如果该模型是过大(例如,超过5%),大脑可能死后MRI和/或切片中移动。另外重要的一点是要适应脑模型的设计,使得小脑正确放置在3D打印对象的内部。当小脑已尸检脑提取期间被损坏这可以是特别具有挑战性的。
当打印脑切片机和保持器,三维打印机的类型,也必须仔细地选择。一些多喷墨打印机使用烘箱移除支撑材料需要后处理。而这些打印机可以产生是水密和相对比桌面熔融沉积成型(FDM)打印机更耐用的物体,在加热过程以除去载体可以稍微翘曲的框,产生刀片的间隙是不完全垂直于大脑轮廓。
大脑切片的过程是另一个关键的一步。之前切割日Ë全脑入板,以确保大脑紧紧地坐在大脑切片机里面是非常重要的:应该有,当施加到大脑轻微的压力没有运动。这将有可能使叶片通过脑切在由研究者设定的精确位置。连续的,平衡的压力应在切割时被应用到刀片夹。取决于叶片的锐度,而组织的刚性,有轻微的横向切割运动可以是用于保持平坦切断面是有利的。
石蜡嵌入处理也可以是MRI和组织学之间的未对准的一个来源。如果该组织板坯不平坦靠在盒在嵌入处理期间坐在,会有切片机的切割平面和板坯的表面的地方之间的倾斜。这将需要切割不可用部分,以找到其中所有的组织暴露的平面。以校正倾斜单程通过改变观察面的角度对高各向同性分辨率死后MRI检查。然而,这几乎是不可能的上通常具有各向异性的分辨率(通常厚冠状切片)收购体内MRI执行。
最后,组织能,以及在幻灯片(折叠,龟裂,皱纹)的工作经历在福尔马林固定期限和石蜡包埋(收缩)有些变形。一些这些变形可以通过将在水浴4-5微米的部分转印到幻灯片之前进行校正。其它变形可通过对死后MRI图像进行组织学数字化图像的可变形图像配准来部分地解决。然而,仔细和熟练的实践尽量减少变形是最有效的方法来匹配MRI卷组织切片。
总之,这里介绍的方法使INVestigators准确评估MRI表现的基本病理。更一般地,它是用于识别和/或验证对调查研究针对特定的病理过程,如炎症或髓鞘新颖的MRI标记物有希望的方法。
披露声明
The authors declare that they have no competing financial interests.
致谢
The Intramural Research Program of NINDS supported this study. We thank the NIH Functional Magnetic Resonance Imaging Facility. We thank Jennifer Lefeuvre and Cecil Chern-Chyi Yen for assistance with postmortem MRI acquisition. We thank John Ostuni and the Section on Instrumentation Core Facility for assistance with 3D printing. Figure 1 of this work used snapshots from MeshLab, a tool developed with the support of the 3D-CoForm project.
材料
| Name | Company | Catalog Number | Comments |
| 7T/30cm USR AVIII Bruker MRI | Bruker Biospin | ||
| 38 mm Bruker Biospin volume coil | Bruker Biospin | ||
| Fomblin | Solvay Solexis | ||
| 50 ml Falcon Centrifuge Tubes, Polypropylene, Sterile | Corning | 21008-951 | |
| Fisherbrand Gauze Sponges | Fisher Scientrific | 13-761-52 | |
| Parafilm M All-Purpose Laboratory Film | Bemis | ||
| Leica RM2235 rotary microtome | Leica | ||
| Leica Disposable Blades, low profile (819) | Leica | ||
| Cresyl Violet Acetate, 0.1% Aqueous | Electron Microscopy Sciences | 26089-01 | |
| Luxol Fast Blue, 0.1% in 95% Alcohol | Electron Microscopy Sciences | 26056-15 | |
| ETOH | |||
| Ultimaker 2 Extended | Ultimaker | ||
| .75 kg Official Ultimaker Branded PLA Filament, 2.85 mm, Silver Metallic | Ultimaker | ||
| Axio Observer.Z1 | Zeiss | ||
| Zen 2 (Blue Edition) | Zeiss | ||
| Netfabb Professional 5.0.1 | Netfabb | http://www.netfabb.com/professional.php | |
| Meshmixer 10.9.332 | Autodesk | http://www.meshmixer.com/download.html | |
| Mipav 7.2 | NIH CIT | http://mipav.cit.nih.edu | |
| Cura | Ultimaker | https://ultimaker.com/en/products/cura-software |
参考文献
- Evans, A. C., Frank, J. A., Antel, J., Miller, D. H. The role of MRI in clinical trials of multiple sclerosis: comparison of image processing techniques. Ann Neurol. 41 (1), 125-132 (1997).
- 't Hart, B. A., van Kooyk, Y., Geurts, J. J. G., Gran, B. The primate autoimmune encephalomyelitis model; a bridge between mouse and man. Ann Clin Transl Neurol. 2 (5), 581-593 (2015).
- Ontaneda, D., Hyland, M., Cohen, J. A. Multiple sclerosis: new insights in pathogenesis and novel therapeutics. Annu Rev Med. 63, 389-404 (2012).
- Guy, J. R., Sati, P., Leibovitch, E., Jacobson, S., Silva, A. C., Reich, D. S. Custom fit 3D-printed brain holders for comparison of histology with MRI in marmosets. J Neurosci Methods. 257, 55-63 (2016).
- Breen, M. S., Lazebnik, R. S., Wilson, D. L. Three-dimensional registration of magnetic resonance image data to histological sections with model-based evaluation. Ann Biomed Eng. 33 (8), 1100-1112 (2005).
- Dauguet, J., et al. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. J Neurosci Methods. 164 (1), 191-204 (2007).
- McGrath, D. M., Vlad, R. M., Foltz, W. D., Brock, K. K. Technical note: fiducial markers for correlation of whole-specimen histopathology with MR imaging at 7 tesla. Med Phys. 37, 2321-2328 (2010).
- Schormann, T., Zilles, K. Three-Dimensional linear and nonlinear transformations: An integration of light microscopical and MRI data. Hum Brain Mapp. 6, 339-347 (1998).
- Jiang, L., et al. Combined MR, fluorescence and histology imaging strategy in a human breast tumor xenograft model. NMR Biomed. 26 (3), 285-298 (2013).
- Absinta, M., et al. Postmortem magnetic resonance imaging to guide the pathologic cut: individualized, 3-dimensionally printed cutting boxes for fixed brains. J Neuropathol Exp Neurol. 73 (8), 780-788 (2014).
- Gaitán, M. I., et al. Perivenular brain lesions in a primate multiple sclerosis model at 7-tesla magnetic resonance imaging. Mult Scler. 20 (1), 64-71 (2014).
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。