需要订阅 JoVE 才能查看此. 登录或开始免费试用。
Method Article
高通量识别对伪单鱼的电阻syringae pv。番茄中的番茄使用苗子洪水测定
Erratum Notice
摘要
幼苗洪水测定有助于快速筛选野生番茄加入,以抵抗伪莫纳斯西林加细菌。这种测定与幼苗细菌生长测定结合使用,有助于进一步描述细菌的潜在抗药性,并可用于筛选种群图以确定抗药性的遗传基础。
摘要
番茄是一种农学上重要的作物,可以感染伪莫纳斯西林加,一种革兰氏阴性细菌,导致细菌斑点病。番茄-P. 西林格pv。番茄病系广泛用于解剖植物先天反应和抗病的遗传基础。几十年来,通过将从索拉伦皮皮内利菌引入培养番茄的Pto/Prf基因簇成功地管理疾病,而P.syringae的种族1菌株已经进化,以克服托/普夫基因簇赋予的阻力,并在全世界发生。
野生番茄品种是病原体识别中自然多样性的重要蓄水池,因为它们在不同环境、不同病原体压力下进化。在野生番茄抗病的典型屏幕中,使用成年植物,这可以限制植物的数量,这些植物可以筛选,因为它们的生长时间延长,生长空间要求更大。我们开发了一种筛选10天大番茄幼苗的抗性方法,将植物生长时间和生长室空间降至最低,使植物快速周转,并允许测试大样本尺寸。生存或死亡的幼苗结果可视为离散表型或受洪水后存活幼苗新生长量定义的阻力尺度。该方法经过优化,筛选了10天大的番茄幼苗,以抵抗两个P.syringae菌株,并可轻松适应其他P.syringae菌株。
引言
伪莫纳斯西林加是一种葛兰阴性致病菌,可感染多种植物宿主。细菌通过气孔或物理伤口进入宿主植物,并在凋亡1中增殖。植物已经进化出一种双层免疫反应,以防止细菌病原体感染。第一级发生在植物细胞表面,植物细胞膜上的模式识别受体在称为PAMP触发免疫(PTI)2的过程中感知高度保守的病原体相关分子模式(PAMPs)。在这个过程中,宿主植物加强防御反应途径,包括将钙质沉积到细胞壁上,关闭气孔,生产活性氧物种,以及诱导发病机制相关基因。
细菌可以通过利用III型分泌系统将蛋白质(称为效应器)直接输送到植物细胞3中,从而克服PTI。效应蛋白通常针对PTI的成分和促进病原体毒性4。第二层植物免疫在识别效应蛋白后发生在植物细胞内。这种识别依赖于抗药性基因,这些基因编码核苷酸结合位点富含白氨酸的重复含有受体(NLRs)。NLR 能够直接识别效应者或识别其在毒力目标或诱饵5上的活动。然后,它们触发一种称为效应器触发免疫(ETI)的过程的继发免疫反应,该反应通常与超敏反应(HR)相关,这是感染6地点的局部细胞死亡形式。与ETI相关的基因对基因电阻不同,植物可以表现出定量的局部阻力,这依赖于多个基因7的贡献。
P. 西林格加pv.番茄是番茄上细菌斑点的因果剂,是一个长期的农业问题.场上的主要菌株通常是Pst比赛0菌株,表示或两种III型效应器AvrPto和AvrPtoB。DC3000(PstDC3000) 是一种具有代表性的种族 0 菌株和一种模型病原体,可引起番茄中的细菌斑点。为了对抗细菌斑点病,育种者将Pto [P. syringae pv. 番茄]/Prf =Pto电阻和芬西翁敏感性] 基因簇从野生番茄物种索兰皮皮内利菌进入现代品种8,9.托基因编码的丝氨酸-threonine蛋白激酶,与Prf NLR一起,通过识别效应器AvrPto和AvrPtoB10,11,12,13,14,赋予对PstDC3000的电阻。然而,这种抵抗对新出现的比赛1株是无效的,允许他们快速和侵略性蔓延,近年来15,16。比赛1菌株逃避Pto/Prf集群的识别,因为AvrPto在这些菌株中丢失或突变,而AvrPtoB似乎累积最少15,17,18。
野生番茄种群是Pst抗药性自然变化的重要储藏库,以前曾用于识别潜在的抗性位点19、20、21。然而,目前用于病原体耐药性的屏幕使用4-5周大的成年植物20,21。因此,它们受生长时间、生长室空间和相对较小的样品尺寸的限制。为了解决传统方法的局限性,我们利用10天大的番茄苗22开发了高通量番茄P.syringae抗性测定。与使用成人工厂(即缩短生长时间、减少空间要求和提高吞吐量)比较,此方法具有几个优点。此外,我们已经证明,这种方法忠实地概括了在成年植物22中观察到的抗病现象。
在该协议中描述的幼苗洪水测定中,番茄幼苗生长在无菌的Murashige和Skoog(MS)介质的培养皿上,为期10天,然后被含有感兴趣的细菌和表面活性剂的细菌淹没。洪水过后,可以通过细菌生长测定定量评估幼苗的抗病能力。此外,幼苗的生存或死亡可以作为离散的抵抗或疾病表型7-14天后洪水。这种方法提供了一个高通量的替代方案,用于筛选大量的野生番茄加入,以抵抗Pst种族1菌株,如Pst菌株T1(PstT1),并可以很容易地适应其他细菌菌株的兴趣。
Access restricted. Please log in or start a trial to view this content.
研究方案
1. 生物安全柜的制备和使用
- 用70%乙醇擦拭生物安全柜。
- 关闭窗框,打开生物安全柜中的紫外线 15 分钟。
- 15分钟后,关闭生物安全柜中的紫外线。提起窗框并打开鼓风机 15 分钟。
- 在将物品放入消毒柜之前,用 70% 乙醇擦拭生物安全柜中使用的所有物品。
- 在生物安全柜中工作之前,用 70% 乙醇清洁手套或徒手。
- 在生物安全柜的中心工作,远离鼓风机。
- 使用未开封的无菌瓶 10 mM MgCl2和超纯 H2O 进行实验。将瓶子放入生物安全柜中,只打开消毒的生物安全柜,而不是放在台面上。
- 使用专用玻璃移液器和移液器提示在消毒的生物安全柜中工作。确保这些仅在生物安全柜中打开,切勿在台式上打开。
- 使用生物安全柜后,高压灭菌器将所有废物(漂白废物除外)和用70%乙醇擦拭表面。
2. 植物介质的制备
- 在超纯H2O中称重和溶解0.5倍MS基底盐。 称量0.8%的巴多加,然后加入溶解的0.5倍MS。
- 高压灭菌,让介质在浇注或移液前在50°C水浴中冷却1小时。
- 为确保板材不过度填充,将聚苯乙烯一次性无菌 100 x 25 mm 板标记为 40 mL 的填充水平。将介质倒入 100 x 25 mm 无菌板中,放入消毒的生物安全柜中。
3. 植物材料和生长条件的制备
- 将番茄种子放入2.2 mL微离心管中,加入2.0 mL的50%漂白液。
- 在摇杆上摇动管子 25 分钟。
- 25分钟后,从摇臂中取出种子,用无菌生物安全柜中的移液器去除漂白液。确保所有漂白剂都已去除。
- 加入2 mL无菌超纯H2O洗涤种子。反转管 5 倍。
- 用移液器从管子中取出液体。
- 重复步骤 3.3×3.5,将种子洗净 4 倍以上。
- 加入2 mL的无菌超纯H2O,并将种子倒入一个空无菌培养皿中。
- 火焰力在乙醇中,并允许冷却之前转移和均匀间距的种子在100 x 25毫米板包含0.5x MS + 0.8%的agar介质。
- 将 5⁄7 种子在一个板的中间线中移植,用手术胶带(1.25 厘米 x 9.1 米)密封板的边缘。
- 在黑暗中将灭菌种子在4°C下分层至少3天,以同步发芽。确保盘子平和朝上堆叠,使种子不会在盘子上移动。
- 垂直定向板,使根沿板表面向下生长,当移至生长室时,种子线水平方向。
注:将生长室设置为 22°C,在 200×220 μE 米-2 s-1和 8 小时的黑暗光强度下提供 16 h 的光。 - 在洪水发生之前,在生长室中生长10天,此时幼苗通常充分出现并扩大的木叶和新兴的第一真叶(图1)。

图1:典型10天大番茄苗的发育阶段。里奥格兰德-波托R番茄种子经过消毒、镀层和分层,在黑暗中在4°C下至少3天。幼苗在22°C下在0.5x MS板上生长10天,然后被淹没。通常,在10天,粘落完全扩大,第一个真正的叶子开始出现。请点击此处查看此图形的较大版本。
4. 准备国王B23(KB)介质
- 用 500 mL 超纯 H2O 填充烧杯,并在搅拌盘上搅拌。
- 将20克巴托肽、1.5克无水性K2HPO4和12.5mL甘油完全溶解在超纯H2O的烧杯中。
- 将溶解的混合物倒入 1 L 精化气缸中,使用超纯 H2O 将最终体积提高 1 L。
- 将汤放回烧杯中搅拌,直到混合。
- 将 7.5 克的百藤 agar 称重到两个 500 mL 玻璃瓶中,并从步骤 4.4 中加入 500 mL KB 汤。高压灭菌 20 分钟。
- 从高压灭菌器中取出瓶子,轻轻旋转以分配安格。
- 将瓶子转移到50°C水浴1小时。
- 1小时后,将瓶子转移到生物安全柜,在无菌条件下,加入1,600μL的无菌1 M MgSO4,并适当抗生素到介质中。
注:对于抗利福平耐菌株PstDC3000和PstT1,使用溶解在二甲基酰胺中的利福平,最终浓度为50微克/米。使用溶解在乙醇中的环己胺,最终浓度为50微克/mL,以防止菌在板上生长。 - 轻轻旋转介质以混合,然后倒入板的底部。
- 在4°C倒存之前,让板至少1小时凝固。
5. 维持细菌菌株和培养条件
- 将单菌群中的甘油储存为1 mL饱和细菌培养物,在-80°C时保持333μL的无菌80%甘油。
- 用适当的抗生素将糖醇储存到KB agar上的修补细菌(即PstT1) 到KB agar上(第4节)。
- 让细菌在28°C下恢复2天,然后用扁平无菌牙签将新鲜细菌条纹到选择性KB Agar上。
- 使用扁平无菌牙签将新鲜细菌从甘油库存中划到适当的选择性KB agar上。
注:确保修补的甘油库存不超过2周。 - 对于PstDC3000,在洪水实验中使用细菌之前,在 28 °C 下孵育 KB 板 24 小时。
- 对于PstT1,在洪水实验中使用细菌之前,在 28 °C 下孵育 KB 板 48 小时。
6.准备PstT1接种
- 无菌地将无菌10 mM MgCl2中的细菌重新悬浮到光学密度为600nm(OD600)为0.1,或大约5 x 107菌落形成单元(CFU)/mL)。
- 使用无菌 10 mM MgCl2溶液在生物安全柜中执行串行稀释。对于PstT1,使用分光光度计使起始浓度为 OD600 = 0.1 的接种。
- 对于PstT1,从 OD600 = 0.1 的初始重新悬浮器进行 1/10 稀释,以获得 OD600 = 0.01 浓度的串行稀释。
- 使用 OD600 = 步骤 6.3 的 0.01 处的串行稀释,进行 3/4 稀释以获得最终 OD600 = 0.0075。
- 在10 mM MgCl2中对非离子有机硅表面活性剂共聚物C13H34O4Si3(即表面活性剂)进行1/10稀释,将1/10的浮层稀释剂加入最后一个串行稀释(OD600 = 0.0075),最终浓度为0.015%,混合到混合井。
7.准备PstDC3000接种
- 无菌地将无菌10 mM MgCl2中的细菌悬浮到光学密度为600nm(OD600)0.1(约5 x 107 CFU/mL)。
- 使用无菌 10 mM MgCl2溶液在生物安全柜中执行串行稀释。对于PstDC3000,使用分光光度计使起始浓度为 OD600 ± 0.1 的接种。
- 对于PstDC3000,从 OD600 = 0.1 的初始重新悬浮进行 1/10 稀释,以获得 OD600 = 0.01 浓度的串行稀释。
- 使用第 3 步 OD600 = 0.01 处的串行稀释,进行 1/2 稀释以获得最终 OD600 = 0.005。
- 在 10 mM MgCl2和 15 s 的涡旋中使表面活性剂稀释 1/10。 将 1/10 库存的表面活性剂添加到最后一个串行稀释(OD600 = 0.005)中,最终浓度为 0.015%,旋转均匀混合。
8. 番茄育苗洪水法
- 将10天大幼苗从生长室中拿出来,放入生物安全柜,为防洪板做准备。
- 从两个盘子中取出手术胶带。
- 设置 3 分钟的计时器,测量 6 mL 的最终接种器(PstT1 OD600 = 0.0075 [第 6 节] 或PstDC3000 OD600 = 0.005 [第 7 节]),并将 6 mL 的鼻孔与 10 天幼苗一起转移到每个板。
- 用无菌移液器尖端轻轻将幼苗向下推入接种。启动计时器。
- 每只手拿着一个盘子。向下倾斜板的前面,以积累幼苗,并主要淹没幼苗的树苗和叶子。
- 将侧面侧向 5⁄7x,然后将板倒回以盖住根和整个板。
- 再次向下倾斜板,以淹没粘结的孔和叶子,并重复总共3分钟。
- 将内孔从板上倒出,将板放在平坦的表面上,然后第二次倒出任何残留的内孔。
- 用手术胶带重新包装板,并重复步骤 8.2_8.8,用于任何剩余的板。
- 所有板被水淹后,重新孵育生长室中的板(参见步骤 3.11 注)。
- PstDC3000 的 7-10 天后的表型,PstT1 的 10-14 天(第 11 节)。如果进行细菌生长测定,在4天(第9节和第10节)后收集叶组织,然后进行表型(第11节)。或者,对单独的植物组进行模型分析和细菌生长测定。
9. 细菌生长测定的粘剂表面灭菌
- 洪水泛滥四天后,在生长室重新孵育幼苗(第8节),用番茄苗子从生长室中取出盘子。
- 编号板底外侧的单个幼苗,其中幼苗附着在每个基因型的板上。
- 标签无菌 1.5 mL 微离心管与单独的幼苗编号,并使用干净的钳子将一个 3 mm 无菌硼硅酸盐珠滴到每个管与珠子搅拌器一起使用。(请参阅步骤 10.1 中的注释。
- 移液器 200 μL 10 mM MgCl2进入每个管和闭管。
- 准备70%乙醇,将100 mL倒入清洁的烧杯中。将 100 mL 无菌超纯 H2O 倒入单独的清洁烧杯中。
- 使用乙醇锯齿尖清洁不锈钢直细点钳。稍微打开板,以便用干净的钳子无菌去除一个粘结剂。
- 捏在焦质的底面的小菜,取出一片叶子,用70%乙醇放入烧杯中,进行10s的表面消毒。
- 将卡蒂莱顿放在纸巾上,用精致的科学湿巾擦干。
- 表面灭菌和印迹后单独称重每个粘液,并记录重量。
- 将卡蒂莱顿放入先前准备的1.5mL微离心管(从步骤9.3和9.4起),标有相应的基因型和单个编号。
- 用无菌胶带重新密封板,重新孵育生长室中的幼苗(参见步骤 3.11 注)。
10. 细菌生长测定
- 使用步骤 9.10 中的样本,使用 10 mM MgCl2中的珠子搅拌器将组织均匀化 1⁄2 分钟。如果组织没有充分浸渍,再次均匀。
注:许多制造商生产珠子搅拌器均质器。珠子的数量和类型,以及同质化时间和速度(如果是可编程的)应针对每种类型的均质器进行优化。确保样品在均匀化期间不会过热。 - 从步骤 10.1 到每个含有浸渍组织的管中加入 800 μL 的 10 mM MgCl2,然后反转几次混合。
- 使用多通道移液器(图2A),在96井板(10 0、10-1、10-2、10-3、10-4、10-5)中,为10 mM MgCl2中的每个样品准备序列稀释。
- 从每个稀释系列移液器5 μL使用多通道移液器到KB加盘(150毫米x15毫米)与环氧西米德和适当的选择细菌感兴趣的菌株(参见步骤4.8注)。让盘子完全干燥。
- 在28°C下将板倒置36小时,然后用解剖显微镜将板上的菌落可视化(图2B),以确定菌落是否足够大以计数。
注:如果菌落不够大,则重新孵化盘子,每隔几小时重新检查一次菌落的大小。通常,菌落在孵育后可计数[36~48小时]

图2:苗子细菌生长测定的序列稀释。(A) 受感染植物的梅化叶组织在群落计数前被稀释。稀释在 96 孔板中执行(未稀释 100)。通常,稀释由 10-1到 10-5。(B) 为细菌菌群计数的电镀稀释物。稀释系列每一柱共镀5μL,从最稀稀释到最集中。菌落完全干燥后,在28°C孵育36~48小时。 菌落在10倍解剖显微镜下计数。请点击此处查看此图形的较大版本。
- 在菌落合并之前,在解剖显微镜下计数(图2B)。从少于 100 个菌落的稀释系列板中计数菌落。
- 获得菌落计数(图2B)后,将幼苗的计数标准化为0.01克组织,并转换为原体细菌生长(表1)。
注:一个货币制造商-PtoS cotyledon的平均质量是0.01克,是经验确定22。
| 基因型1列 A | 组织重量 (g) 列 B | 点列 C 中的殖民地的 + | 点2列 D 的稀释系数 | 调整的殖民地 3 列 E 的 # | 串行稀释柱 F 的稀释系数 | 殖民地 G 柱总数 (cfu/0.01 g)4 | 殖民地的平均数 (cfu/0.01 g) 柱 H | 平均日志增长(cfu/0.01 g(日志10)列 I |
| 示例 1 | 0.004 克 | 10 | 200 | 计算为: (C2 x 0.01 g) / B2 = 25 | 1000 | 计算为: (D2 x E2 x F2) = 5000000 | 样本 1 到最后一个样本的平均值: (即平均 G1:G3) = 7000000 | 平均日志。日志(H2) = 6.85 |
| 示例 2 | 0.003 克 | 15 | 200 | 50 | 1000 | 10000000 | ||
| 示例 3 | 0.002 克 | 6 | 200 | 30 | 1000 | 6000000 | ||
| 1显示 3 个样本的数据 | ||||||||
| 2基于电镀 5 μL x 200,用于 1 mL | ||||||||
| 3科蒂莱顿太小,不能核心,因此根据一个MoneyMaker-PtoS cotyledon的平均质量将菌体计数标准化为0.01克组织(未显示的数据) | ||||||||
| 4根据镀体积调整每 mL | ||||||||
表1:幼苗细菌生长测定的样本计算。样本计算演示了如何使细菌计数正常化并确定原细菌生长。
- 对于野生加入和其他具有复杂遗传背景的系,如第11节所述,将单个幼苗的细菌生长水平与其表型相关联。
11. 抵抗的平地
- 从生长室和表型单个幼苗中取出板,在7-14天后死亡(因病)或存活(由于耐药性)。
- 在洪水接种后7-10天,感染PstDC3000等高毒性菌株的表型植物。
- 在洪水接种后10-14天感染PstT1的表型植物。
- 根据观察到的电阻表型范围确定评分系统。记录品种、等源线和野生加入的二元表型,具有一致、强到中间电阻的表型(图4A,4B)。
- 如果幼苗在镶势的时间范围内显示来自尖顶的幼苗的新生长,则将其视为生存。如果幼苗有一个棕色的虫绿,并且没有新的绿色植物生长,把它算作死亡(图3)。
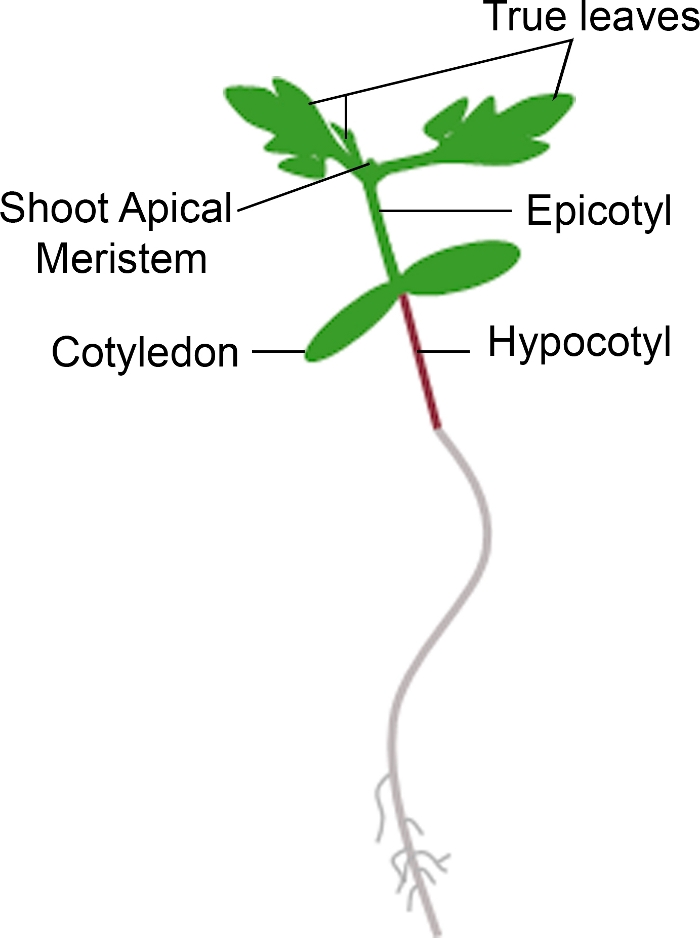
图3:番茄苗的原理图表示。描绘了番茄幼苗的不同部分,包括下科基、科蒂林、史诗、射虫和真叶。请点击此处查看此图形的较大版本。
- 记录具有广泛抗药性表型的种群(图4C)的疾病谱上的现象类型,如F2映射种群。
- 仔细监测幼苗是否出现疾病症状和死亡,以确定适当的表皮窗口。
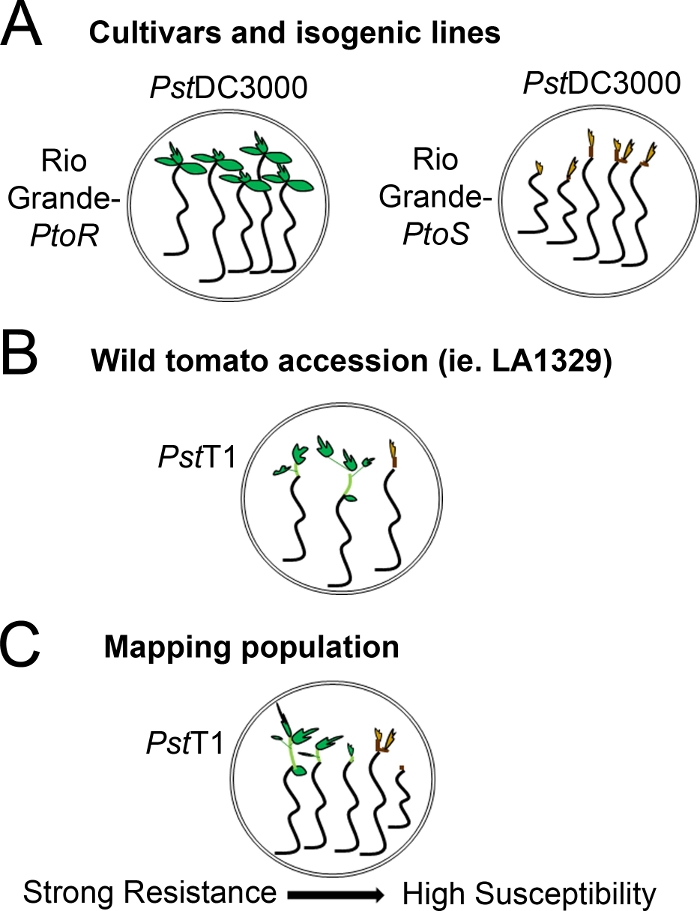
图4:不同遗传背景下幼苗阻力和死亡的预期表型的原理图表示。(A) 里奥格兰德-普托R和近等源性栽培的幼苗在洪水泛滥 7 天后显示PstDC3000 (OD600 × 0.005) = 0.015% 表面活性剂。里奥格兰德-普托R表现出一致的耐药性,而里奥格兰德-PtoS表现出对PstDC3000感染的一致易感性。这些线产生离散和二进制表型。(B) 野生加入的幼苗,如Solanum neorickii LA1329,在PstT1 (OD600 = 0.0075) = 0.015% 表面活性剂泛滥 10 天后显示。幼苗显示表型变异性,但被记录为二元表型。型比变异量和拟合法法(二进制电阻或电阻谱)将取决于所测试的特定加入。(C) 绘制通过穿越野生加入到易感品种生成的种群,在F2隔离种群中可能显示更广泛的表型。在这种情况下,在光谱上记录幼苗表型可能是最合适的。从测绘种群中高度易受影响的幼苗可能早在第7天就被表型为死亡,当被PstT1淹没时,通常表现出棕色的幼虫,不向很少延伸的史诗,也没有新的绿色植物生长。易感幼苗的幼苗可能保持绿色或非常浅棕色的时间,并且可能有一些延长的史诗和很少的植物生长,这变成棕色和逮捕第10天。根据第14天新增和持续植物生长量,可以表型的抗性。然后,根据上述表型将幼苗分组到不同类别的阻力,如弱、中或强阻力。请点击此处查看此图形的较大版本。
Access restricted. Please log in or start a trial to view this content.
结果
利用幼苗电阻测定检测培养物和等源系中的PtoR-介导免疫
图 5显示了货币制造商-PtoR和货币制造商 -PtoS品种在PstDC3000 泛滥 7-10 天后的代表性结果。在感染之前,10天大的幼苗完全出现并扩大,并出现第一个真正的叶子。幼苗在光学密度为0.005~ 0.015%的表面活性剂中,以负控制(未显示数据)和PstDC3000被淹没。幼苗在洪水?...
Access restricted. Please log in or start a trial to view this content.
讨论
介绍了一种使用PstDC3000或PstT1优化的洪水接种方案,以检测番茄幼苗中对这些细菌菌株的抗药性。在幼苗阻力测定中,有几个关键参数,包括细菌浓度和表面活性剂浓度,这是经验确定22。对于PstDC3000,光学密度进行了优化,在含有Pto/Prf簇的耐药品种上实现完全存活,并在缺乏Pto/Prf群集22的易感品种上完全死亡。对于PstT1...
Access restricted. Please log in or start a trial to view this content.
披露声明
作者没有什么可透露的。
致谢
我们感谢杰米·卡尔玛测试媒体量对疾病或耐药性结果的影响。我们感谢刘易斯实验室的Maöl Baudin博士和卡尔·谢伯博士对手稿提出建设性意见和建议。Lewis实验室植物免疫研究得到了美国农业部ARS 2030-21000-046-00D和2030-21000-050-00D(JDL)和NSF生物科学局IOS-1557661(JDL)的支持。
Access restricted. Please log in or start a trial to view this content.
材料
| Name | Company | Catalog Number | Comments |
| 3M Tape Micropore 1/2" x 10 YD CS 240 (1.25 cm x 9.1 m) | VWR International | 56222-182 | |
| 3mm borosilicate glass beads | Friedrich & Dimmock | GB3000B | |
| Bacto peptone | BD | 211677 | |
| Bacto agar | BD | 214010 | |
| Biophotometer Plus | Eppendorf | E952000006 | |
| Biosafety cabinet, class II type A2 | |||
| BRAND Disposable Plastic Cuvettes, Polystyrene | VWR International | 47744-642 | |
| Chenille Kraft Flat Wood Toothpicks | VWR International | 500029-808 | |
| cycloheximide | Research Products International | C81040-5.0 | |
| Dibasic potassium phosphate anhydrous, ACS grade | Fisher Scientific | P288-500 | |
| Dimethylformamide | |||
| Dissecting microscope (Magnification of at least 10x) | |||
| Ethanol - 190 Proof | |||
| Falcon polystyrene 96 well microplates, flat-bottom | Fisher Scientific | 08-772-3 | |
| Glass Alcohol Burner Wick | Fisher Scientific | S41898A / No. W-125 | |
| Glass Alcohol Burners | Fisher Scientific | S41898 / No. BO125 | |
| Glycerol ACS reagent | VWR International | EMGX0185-5 | |
| Kimberly-Clark™ Kimtech Science™ Kimwipes™ Delicate Task Wipers | Fisher Scientific | 06-666-A | |
| Magnesium chloride, ACS grade | VWR International | 97061-356 | |
| Magnesium sulfate heptahydrate, ACS grade | VWR International | 97062-130 | |
| Microcentrifuge tubes, 1.5 mL | |||
| Microcentrifuge tubes, 2.2 mL | |||
| Mini Beadbeater-96, 115 volt | Bio Spec Products Inc. | 1001 | |
| Murashige & Skoog, Basal Salts | Caisson Laboratories, Inc. | MSP01-50LT | |
| Pipet-Lite XLS LTS 8-CH Pipet 20-200uL | Rainin | L8-200XLS | |
| Pipet-Lite XLS LTS 8-CH Pipet 2-20uL | Rainin | L8-20XLS | |
| Polystyrene 100mm x 25mm sterile petri dish | VWR International | 89107-632 | |
| Polystyrene 150mm x 15mm sterile petri dish | Fisher Scientific | FB08-757-14 | |
| Polystyrene 150x15mm sterile petri dish | Fisher Scientific | 08-757-148 | |
| Pure Bright Germicidal Ultra Bleach 5.7% Available Chlorine (defined as 100% bleach) | Staples | 1013131 | |
| Rifampicin | Gold Biotechnology | R-120-25 | |
| Silwet L-77 (non-ionic organosilicone surfactant co-polymer C13H34O4Si3 surfactant) | Fisher Scientific | NCO138454 | |
| Tips LTS 20 μL 960/10 GPS-L10 | Rainin | 17005091 | |
| Tips LTS 250 μL 960/10 GPS-L250 | Rainin | 17005093 | |
| VWR dissecting forceps fine tip, 4.5" | VWR International | 82027-386 |
参考文献
- Underwood, W., Melotto, M., He, S. Y. Role of plant stomata in bacterial invasion. Cell Microbiology. 9 (7), 1621-1629 (2007).
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Current Opinion in Plant Biology. 12 (4), 414-420 (2009).
- Galan, J. E., Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 444 (7119), 567-573 (2006).
- Lewis, J. D., Desveaux, D., Guttman, D. S. The targeting of plant cellular systems by injected type III effector proteins. Seminars in Cell and Developmental Biology. 20 (9), 1055-1063 (2009).
- Schreiber, K. J., Baudin, M., Hassan, J. A., Lewis, J. D. Die another day: molecular mechanisms of effector-triggered immunity elicited by type III secreted effector proteins. Seminars in Cell and Developmental Biology. 56, 124-133 (2016).
- Heath, M. C. Hypersensitive response-related death. Plant Molecular Biology. 44 (3), 321-334 (2000).
- Boyd, L. A., Ridout, C., O'Sullivan, D. M., Leach, J. E., Leung, H. Plant-pathogen interactions: disease resistance in modern agriculture. Trends in Genetics. 29 (4), 233-240 (2013).
- Pitblado, R. E., MacNeill, B. H. Genetic basis of resistance to Pseudomonas syringae pv. tomato in field tomatoes. Canadian Journal of Plant Pathology. 5 (4), 251-255 (1983).
- Pedley, K. F., Martin, G. B. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annual Reviews of Phytopathology. 41, 215-243 (2003).
- Ronald, P. C., Salmeron, J. M., Carland, F. M., Mehta, A. Y., Staskawicz, B. J. The cloned avirulence gene AvrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. Journal of Bacteriology. 174 (5), 1604-1611 (1992).
- Martin, G. B., et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 262 (5138), 1432-1436 (1993).
- Salmeron, J. M., Barker, S. J., Carland, F. M., Mehta, A. Y., Staskawicz, B. J. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell. 6 (4), 511-520 (1994).
- Salmeron, J. M., et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 86 (1), 123-133 (1996).
- Scofield, S. R., et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 274 (5295), 2063-2065 (1996).
- Kunkeaw, S., Tan, S., Coaker, G. Molecular and evolutionary analyses of Pseudomonas syringae pv. tomato race 1. Molecular Plant-Microbe Interactions. 23 (4), 415-424 (2010).
- Cai, R., et al. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathogens. 7 (8), 1002130(2011).
- Almeida, N. F., et al. A draft genome sequence of Pseudomonas syringae pv. tomato T1 reveals a type III effector repertoire significantly divergent from that of Pseudomonas syringae pv. tomato DC3000. Molecular Plant-Microbe Interactions. 22 (1), 52-62 (2009).
- Lin, N. C., Abramovitch, R. B., Kim, Y. J., Martin, G. B. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Applied and Environmental Microbiology. 72 (1), 702-712 (2006).
- Rose, L. E., Langley, C. H., Bernal, A. J., Michelmore, R. W. Natural variation in the Pto pathogen resistance gene within species of wild tomato (Lycopersicon). I. Functional analysis of Pto alleles. Genetics. 171 (1), 345-357 (2005).
- Thapa, S. P., Miyao, E. M., Davis, R. M., Coaker, G. Identification of QTLs controlling resistance to Pseudomonas syringae pv. tomato race 1 strains from the wild tomato Solanum habrochaites LA1777. Theoretical and Applied Genetics. 128 (4), 681-692 (2015).
- Bao, Z. L., et al. Identification of a candidate gene in Solanum habrochaites for resistance to a race 1 strain of Pseudomonas syringae pv. tomato. Plant Genome. 8 (3), 1-15 (2015).
- Hassan, J. A., Zhou, Y. J., Lewis, J. D. A rapid seedling resistance assay identifies wild tomato lines that are resistant to Pseudomonas syringae pv. tomato race 1. Molecular Plant-Microbe Interactions. 30 (9), 701-709 (2017).
- King, E. O., Ward, M. K., Raney, D. E. Two simple media for the demonstration of pyocyanin and fluorescin. Journal of Laboratory and Clinical Medicine. 44 (2), 301-307 (1954).
- Uppalapati, S. R., et al. Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: phenotypic and gene expression analyses of the virulence function of coronatine. Molecular Plant-Microbe Interactions. 21 (4), 383-395 (2008).
- Bhardwaj, V., Meier, S., Petersen, L. N., Ingle, R. A., Roden, L. C. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One. 6 (10), 26968(2011).
- Lu, H., McClung, C. R., Zhang, C. Tick tock: circadian regulation of plant innate immunity. Annual Review of Phytopathology. 55, 287-311 (2017).
- Wang, W., et al. Timing of plant immune responses by a central circadian regulator. Nature. 470 (7332), 110-114 (2011).
Access restricted. Please log in or start a trial to view this content.
Erratum
Formal Correction: Erratum: High-Throughput Identification of Resistance to Pseudomonas syringae pv. Tomato in Tomato using Seedling Flood Assay
Posted by JoVE Editors on 10/18/2023. Citeable Link.
An erratum was issued for: High-Throughput Identification of Resistance to Pseudomonas syringae pv. Tomato in Tomato using Seedling Flood Assay. The Introduction, Protocol, Representative Results and Discussion sections were updated.
The last paragraph of the Introduction section was updated from:
In the seedling flood assay described in this protocol, tomato seedlings are grown on Petri dishes of sterile Murashige and Skoog (MS) media for 10 days and then are flooded with an inoculum containing the bacteria of interest and a surfactant. Following flooding, seedlings can be quantitatively evaluated for disease resistance via bacterial growth assays. Additionally, seedling survival or death can act as a discrete resistance or disease phenotype 7–14 days after flooding. This approach offers a high-throughput alternative for screening large numbers of wild tomato accessions for resistance to Pst race 1 strains, such as Pst strain T1 (PstT1), and can easily be adapted to other bacterial strains of interest.
to:
In the seedling flood assay described in this protocol, tomato seedlings are grown on Petri dishes of sterile Murashige and Skoog (MS) media for 10 days and then are flooded with an inoculum containing the bacteria of interest and a surfactant. Following flooding, seedlings can be quantitatively evaluated for disease resistance via bacterial growth assays. Additionally, seedling survival or death can act as a discrete resistance or disease phenotype 7–14 days after flooding. This approach offers a high-throughput alternative for screening large numbers of wild tomato accessions for resistance to Pst race 1 strains, such as Pst strain 19 (Pst19), and can easily be adapted to other bacterial strains of interest.
Step 4.8 of the Protocol section was updated from:
- After 1 h, transfer the bottle to the biosafety cabinet and under aseptic conditions, add 1,600 µL of sterile 1 M MgSO4, and appropriate antibiotics to the media.
NOTE: For rifampicin resistant strains PstDC3000 and PstT1, use rifampicin dissolved in dimethylformamide at a final concentration of 50 µg/mL. Use cycloheximide dissolved in ethanol at a final concentration of 50 µg/mL to prevent fungal growth on the plates.
to:
- After 1 h, transfer the bottle to the biosafety cabinet and under aseptic conditions, add 1,600 µL of sterile 1 M MgSO4, and appropriate antibiotics to the media.
NOTE: For rifampicin resistant strains PstDC3000 and Pst19, use rifampicin dissolved in dimethylformamide at a final concentration of 50 µg/mL. Use cycloheximide dissolved in ethanol at a final concentration of 50 µg/mL to prevent fungal growth on the plates.
Step 5.2 of the Protocol section was updated from:
- Patch bacteria (i.e., PstT1) from a glycerol stock onto KB agar with appropriate antibiotics (section 4).
to:
- Patch bacteria (i.e., Pst19) from a glycerol stock onto KB agar with appropriate antibiotics (section 4).
Step 5.6 of the Protocol section was updated from:
- For PstT1, incubate the KB plate at 28 ˚C for 48 h prior to using bacteria in the flood experiment.
to:
- For Pst19, incubate the KB plate at 28 ˚C for 48 h prior to using bacteria in the flood experiment.
Step 6 of the Protocol section was updated from:
6. Preparation of PstT1 inoculum
to
6. Preparation of Pst19 inoculum
Step 6.2 of the Protocol section was updated from:
- Perform serial dilutions using sterile 10 mM MgCl2 solution in the biosafety cabinet. For PstT1, use a spectrophotometer to make inoculum with a starting concentration of OD600 = 0.1.
to:
- Perform serial dilutions using sterile 10 mM MgCl2 solution in the biosafety cabinet. For Pst19, use a spectrophotometer to make inoculum with a starting concentration of OD600 = 0.1.
Step 6.3 of the Protocol section was updated from:
- For PstT1, make a 1/10 dilution from the initial resuspension at OD600 = 0.1 to obtain a serial dilution at a concentration of OD600 = 0.01.
to:
- For Pst19, make a 1/10 dilution from the initial resuspension at OD600 = 0.1 to obtain a serial dilution at a concentration of OD600 = 0.01.
Step 8.3 of the Protocol section was updated from:
- Set a timer for 3 min. Measure 6 mL of final inoculum (PstT1 OD600 = 0.0075 [section 6] or PstDC3000 OD600 = 0.005 [section 7]) and transfer 6 mL of inoculum to each plate with the 10-day-old seedlings.
to:
- Set a timer for 3 min. Measure 6 mL of final inoculum (Pst19 OD600 = 0.0075 [section 6] or PstDC3000 OD600 = 0.005 [section 7]) and transfer 6 mL of inoculum to each plate with the 10-day-old seedlings.
Step 8.11 of the Protocol section was updated from:
- Phenotype after 7–10 days for PstDC3000 or 10–14 days for PstT1 (section 11). If carrying out bacterial growth assays, collect leaf tissue after 4 days (sections 9 and 10) and then phenotype (section 11). Alternatively, perform phenotypic analysis and bacterial growth assays on separate sets of plants.
to:
- Phenotype after 7–10 days for PstDC3000 or 10–14 days for Pst19 (section 11). If carrying out bacterial growth assays, collect leaf tissue after 4 days (sections 9 and 10) and then phenotype (section 11). Alternatively, perform phenotypic analysis and bacterial growth assays on separate sets of plants.
Step 10.7 of the Protocol section was updated from:
- After obtaining colony counts (Figure 2B), normalize the counts to 0.01 g of tissue for seedlings and convert to log bacterial growth (Table 1).
NOTE: The average mass of one Moneymaker-PtoS cotyledon is 0.01 g and is empirically determined22.
| Genotype1 Column A | Tissue Weight (g) Column B | # of Colonies in a spot Column C | Dilution factor for spot2 Column D | Adjusted # of Colonies3 Column E | Dilution factor for serial dilution Column F | Total # of Colonies Column G (cfu/0.01 g)4 | Average # of Colonies (cfu/0.01 g) Column H | Average Log Growth (cfu/0.01 g (log10)) Column I |
| Sample 1 | 0.004 g | 10 | 200 | calculated as: (C2 x 0.01 g) / B2 = 25 | 1000 | calculated as: (D2 x E2 x F2) = 5000000 | average for sample 1 through last sample: (ie. average G1:G3) = 7000000 | log of average ie. log(H2) = 6.85 |
| Sample 2 | 0.003 g | 15 | 200 | 50 | 1000 | 10000000 | ||
| Sample 3 | 0.002 g | 6 | 200 | 30 | 1000 | 6000000 | ||
| 1Data shown for 3 samples | ||||||||
| 2Based on plating 5 µL x 200 for 1 mL | ||||||||
| 3Cotyledons are too small to core so colony counts were normalized to 0.01 g of tissue based on the average mass of one MoneyMaker-PtoS cotyledon (data not shown) | ||||||||
| 4Adjusted per mL based on volume plated | ||||||||
Table 1: Sample calculations for seedling bacterial growth assay. Sample calculations demonstrate how to normalize bacterial counts and determine log bacterial growth.
to:
- After obtaining colony counts (Figure 2B), normalize the counts to 0.1 g of tissue for seedlings and convert to log bacterial growth (Table 1).
NOTE: The average mass of one Moneymaker-PtoS cotyledon is 0.1 g and is empirically determined22.
| Genotype1 Column A | Tissue Weight (g) Column B | # of Colonies in a spot Column C | Dilution factor for spot2 Column D | Adjusted # of Colonies3 Column E | Dilution factor for serial dilution Column F | Total # of Colonies Column G (cfu/0.01 g)4 | Average # of Colonies (cfu/0.01 g) Column H | Average Log Growth (cfu/0.1 g (log10)) Column I |
| Sample 1 | 0.04 g | 10 | 200 | calculated as: (C2 x 0.1 g) / B2 = 25 | 1000 | calculated as: (D2 x E2 x F2) = 5000000 | average for sample 1 through last sample: (ie. average G1:G3) = 7000000 | log of average ie. log(H2) = 6.85 |
| Sample 2 | 0.03 g | 15 | 200 | 50 | 1000 | 10000000 | ||
| Sample 3 | 0.02 g | 6 | 200 | 30 | 1000 | 6000000 | ||
| 1Data shown for 3 samples | ||||||||
| 2Based on plating 5 µL x 200 for 1 mL | ||||||||
| 3Cotyledons are too small to core so colony counts were normalized to 0.1 g of tissue based on the average mass of one MoneyMaker-PtoS cotyledon (data not shown) | ||||||||
| 4Adjusted per mL based on volume plated | ||||||||
Table 1: Sample calculations for seedling bacterial growth assay. Sample calculations demonstrate how to normalize bacterial counts and determine log bacterial growth.
Step 11.3 of the Protocol section was updated from:
- Phenotype plants infected with PstT1 at 10–14 days after flood inoculation.
to:
- Phenotype plants infected with Pst19 at 10–14 days after flood inoculation.
Figure 4 in the Protocol section was updated from:

Figure 4: Schematic representation of expected phenotypes for seedling resistance and death in various genetic backgrounds. (A) Seedlings of Rio Grande-PtoR and the near-isogenic cultivar Rio Grande-PtoS are displayed 7 days after flooding with PstDC3000 (OD600 = 0.005) + 0.015% surfactant. Rio Grande-PtoR displays consistent resistance, and Rio Grande-PtoS displays consistent susceptibility to infection with PstDC3000. These lines give rise to discrete and binary phenotypes. (B) Seedlings of a wild accession, such as Solanum neorickii LA1329, are shown 10 days after flooding with PstT1 (OD600 = 0.0075) + 0.015% surfactant. Seedlings display phenotypic variability but were recorded as binary phenotypes. The amount of phenotypic variability and the method of phenotyping (binary resistance or resistance spectrum) will depend on the particular accession tested. (C) Mapping populations generated by outcrossing wild accessions to susceptible cultivars may display a wider spectrum of phenotypes in F2 segregating populations. In this case, it may be most appropriate to record seedling phenotypes on a spectrum. Highly susceptible seedlings from a mapping population may be phenotyped for death as early as day 7 when flooded with PstT1, and typically show a brown apical meristem, no to very little extension of the epicotyl, and no new, green vegetative growth. The apical meristem of susceptible seedlings may stay green or very light brown for more time, and there may be some extension of the epicotyl and very little vegetative growth, which turns brown and arrests by day 10. Individual seedlings can be phenotyped for resistance based on the amount of new and ongoing vegetative growth by day 14. Seedlings can then be grouped based on the phenotypes described above into different categories of resistance such as weak, medium, or strong resistance. Please click here to view a larger version of this figure.
to:
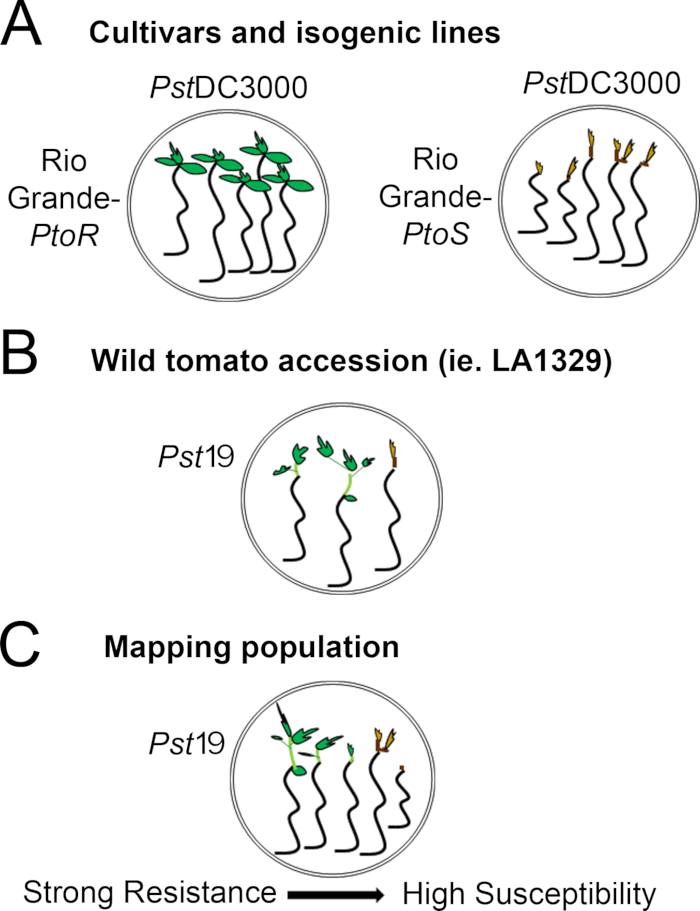
Figure 4: Schematic representation of expected phenotypes for seedling resistance and death in various genetic backgrounds. (A) Seedlings of Rio Grande-PtoR and the near-isogenic cultivar Rio Grande-PtoS are displayed 7 days after flooding with PstDC3000 (OD600 = 0.005) + 0.015% surfactant. Rio Grande-PtoR displays consistent resistance, and Rio Grande-PtoS displays consistent susceptibility to infection with PstDC3000. These lines give rise to discrete and binary phenotypes. (B) Seedlings of a wild accession, such as Solanum neorickii LA1329, are shown 10 days after flooding with Pst19 (OD600 = 0.0075) + 0.015% surfactant. Seedlings display phenotypic variability but were recorded as binary phenotypes. The amount of phenotypic variability and the method of phenotyping (binary resistance or resistance spectrum) will depend on the particular accession tested. (C) Mapping populations generated by outcrossing wild accessions to susceptible cultivars may display a wider spectrum of phenotypes in F2 segregating populations. In this case, it may be most appropriate to record seedling phenotypes on a spectrum. Highly susceptible seedlings from a mapping population may be phenotyped for death as early as day 7 when flooded with Pst19, and typically show a brown apical meristem, no to very little extension of the epicotyl, and no new, green vegetative growth. The apical meristem of susceptible seedlings may stay green or very light brown for more time, and there may be some extension of the epicotyl and very little vegetative growth, which turns brown and arrests by day 10. Individual seedlings can be phenotyped for resistance based on the amount of new and ongoing vegetative growth by day 14. Seedlings can then be grouped based on the phenotypes described above into different categories of resistance such as weak, medium, or strong resistance. Please click here to view a larger version of this figure.
The second paragraph of the Representative Results section was updated from:
Phenotypic screening of wild accessions using the seedling resistance assay
Figure 6 shows representative results for seedlings of susceptible and resistant accessions 10–14 days after flooding with PstT1. Susceptible accessions include RG-PtoR, S. pimpinellifolium LA1375, and S. pimpinellifolium LA1606, and resistant accessions include S. neorickii LA1329. Ten-day-old seedlings were flooded with 10 mM MgCl2 + 0.015% surfactant as a negative control, and PstT1 at an optical density of 0.0075 + 0.015% surfactant. The seedlings were phenotyped at least 10 days after flooding, as PstT1-infected seedlings died more slowly than PstDC3000-infected seedlings. Mock-inoculated seedlings were green, healthy, and actively growing. This control is important to ensure that the accessions are not sensitive to the concentration of surfactant, and to ensure there is no bacterial contamination. Susceptible accessions (Rio Grande-PtoR [n = 7], S. pimpinellifolium LA1375 [n = 7], and S. pimpinellifolium LA1606 [n = 5]) were dead, had brown apical meristems, and lacked new growth 10–14 days after inoculation with PstT1. In contrast, two S. neorickii LA1329 (n = 3) seedlings displayed a high level of new, green growth and survived infection with PstT1 (Figure 6). Three LA1329 seedlings did not germinate. Typically, 5–7 individuals were screened for each accession in a primary screen to determine the prevalence of resistance in the population. When a more genetically complex wild accession, such as LA1329, is flooded with PstT1, the resistance phenotypes display slightly more variability among individual seedlings, compared to Moneymaker-PtoR treated with PstDC3000. However, the resistance phenotypes were usually less variable than those seen in F2 mapping populations. Thus, binary phenotyping criteria was used for LA1329.
to:
Phenotypic screening of wild accessions using the seedling resistance assay
Figure 6 shows representative results for seedlings of susceptible and resistant accessions 10–14 days after flooding with Pst19. Susceptible accessions include RG-PtoR, S. pimpinellifolium LA1375, and S. pimpinellifolium LA1606, and resistant accessions include S. neorickii LA1329. Ten-day-old seedlings were flooded with 10 mM MgCl2 + 0.015% surfactant as a negative control, and Pst19 at an optical density of 0.0075 + 0.015% surfactant. The seedlings were phenotyped at least 10 days after flooding, as Pst19-infected seedlings died more slowly than PstDC3000-infected seedlings. Mock-inoculated seedlings were green, healthy, and actively growing. This control is important to ensure that the accessions are not sensitive to the concentration of surfactant, and to ensure there is no bacterial contamination. Susceptible accessions (Rio Grande-PtoR [n = 7], S. pimpinellifolium LA1375 [n = 7], and S. pimpinellifolium LA1606 [n = 5]) were dead, had brown apical meristems, and lacked new growth 10–14 days after inoculation with Pst19. In contrast, two S. neorickii LA1329 (n = 3) seedlings displayed a high level of new, green growth and survived infection with Pst19 (Figure 6). Three LA1329 seedlings did not germinate. Typically, 5–7 individuals were screened for each accession in a primary screen to determine the prevalence of resistance in the population. When a more genetically complex wild accession, such as LA1329, is flooded with Pst19, the resistance phenotypes display slightly more variability among individual seedlings, compared to Moneymaker-PtoR treated with PstDC3000. However, the resistance phenotypes were usually less variable than those seen in F2 mapping populations. Thus, binary phenotyping criteria was used for LA1329.
Figure 6 in the Representative Results section was updated from:
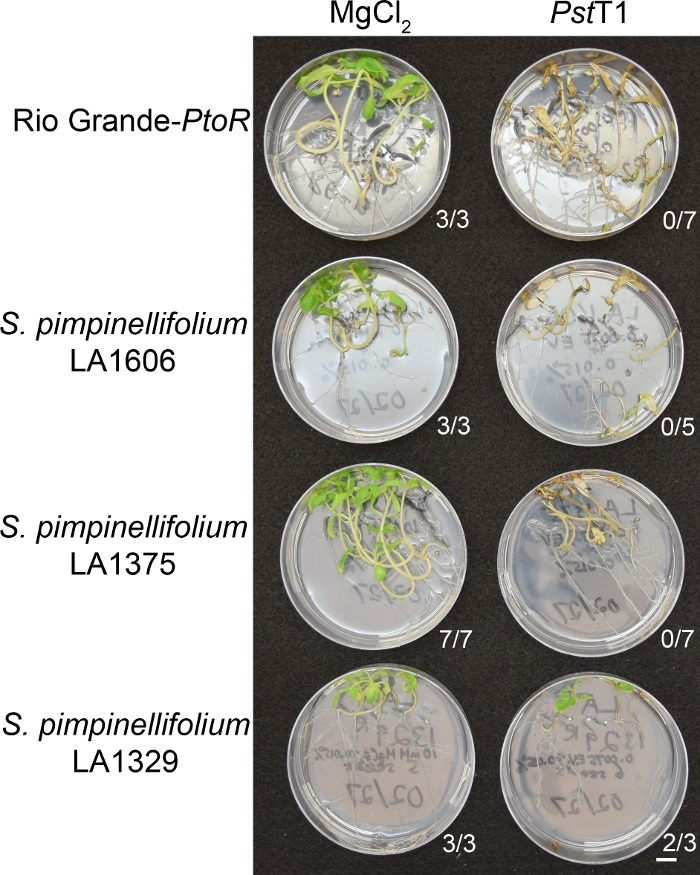
Figure 6: Phenotypic characterization of resistance or disease symptoms 10–14 days post-infection in wild accessions. Rio Grande-PtoR, S. pimpinellifolium LA1606, S. pimpinellifolium LA1375 and S. neorickii LA1329 tomato seedlings were grown on 0.5x MS plates for 10 days, and then flooded with PstT1 (OD600 = 0.0075) + 0.015% surfactant. The number of surviving seedlings for each wild accession out of the total number tested is shown. Scale bar = 1 cm. Please click here to view a larger version of this figure.
to:
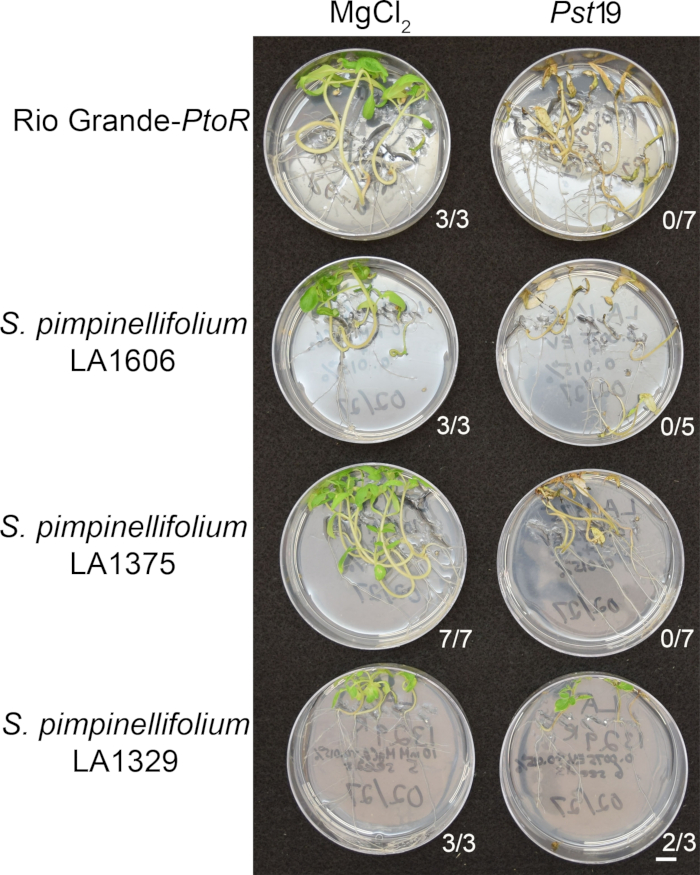
Figure 6: Phenotypic characterization of resistance or disease symptoms 10–14 days post-infection in wild accessions. Rio Grande-PtoR, S. pimpinellifolium LA1606, S. pimpinellifolium LA1375 and S. neorickii LA1329 tomato seedlings were grown on 0.5x MS plates for 10 days, and then flooded with Pst19 (OD600 = 0.0075) + 0.015% surfactant. The number of surviving seedlings for each wild accession out of the total number tested is shown. Scale bar = 1 cm. Please click here to view a larger version of this figure.
The third paragraph of the Representative Results section was updated from:
Quantitative assessment of bacterial growth using the seedling flood assay
To confirm that the observed resistance in LA1329 to PstT1 resulted in lower bacterial growth, bacterial growth assays were carried out in tomato seedlings. The level of PstT1 growth in Moneymaker-PtoS and S. neorickii LA1329 was determined 4 days post-infection. Moneymaker-PtoS is a near-isogenic line with consistent susceptibility among individual seedlings. Wild accessions such as S. neorickii LA1329 are often more genetically complex. LA1329 displays approximately 60% resistance to PstT1 across the population22. Because seedlings may drop their cotyledons after infection, one seedling was grown on each plate to correlate bacterial growth in the harvested cotyledon with overall seedling survival or death as determined phenotypically at least 10 days after flooding. The bacterial counts on day 4 for each seedling were normalized to 0.01 g of tissue and converted to log growth (CFU/0.01 g(log10)). Log growth for phenotypically resistant LA1329 seedlings (LA1329RES) or phenotypically susceptible seedlings (LA1329SUS) were separately pooled and compared to each other and the susceptible cultivar Moneymaker-PtoS. For example, there was a 1.7 log difference in bacterial growth between LA1329RES (log 6.3) and LA1329SUS (log 8.0), and a 1.6 log difference between LA1329RES (log 6.3) and Moneymaker-PtoS (log 7.9) (Figure 7). Therefore, phenotypic resistance correlated with quantitative resistance in the seedling assays.
to:
Quantitative assessment of bacterial growth using the seedling flood assay
To confirm that the observed resistance in LA1329 to Pst19 resulted in lower bacterial growth, bacterial growth assays were carried out in tomato seedlings. The level of Pst19 growth in Moneymaker-PtoS and S. neorickii LA1329 was determined 4 days post-infection. Moneymaker-PtoS is a near-isogenic line with consistent susceptibility among individual seedlings. Wild accessions such as S. neorickii LA1329 are often more genetically complex. LA1329 displays approximately 60% resistance to Pst19 across the population22. Because seedlings may drop their cotyledons after infection, one seedling was grown on each plate to correlate bacterial growth in the harvested cotyledon with overall seedling survival or death as determined phenotypically at least 10 days after flooding. The bacterial counts on day 4 for each seedling were normalized to 0.01 g of tissue and converted to log growth (CFU/0.01 g(log10)). Log growth for phenotypically resistant LA1329 seedlings (LA1329RES) or phenotypically susceptible seedlings (LA1329SUS) were separately pooled and compared to each other and the susceptible cultivar Moneymaker-PtoS. For example, there was a 1.7 log difference in bacterial growth between LA1329RES (log 6.3) and LA1329SUS (log 8.0), and a 1.6 log difference between LA1329RES (log 6.3) and Moneymaker-PtoS (log 7.9) (Figure 7). Therefore, phenotypic resistance correlated with quantitative resistance in the seedling assays.
Figure 7 in the Representative Results section was updated from:
x
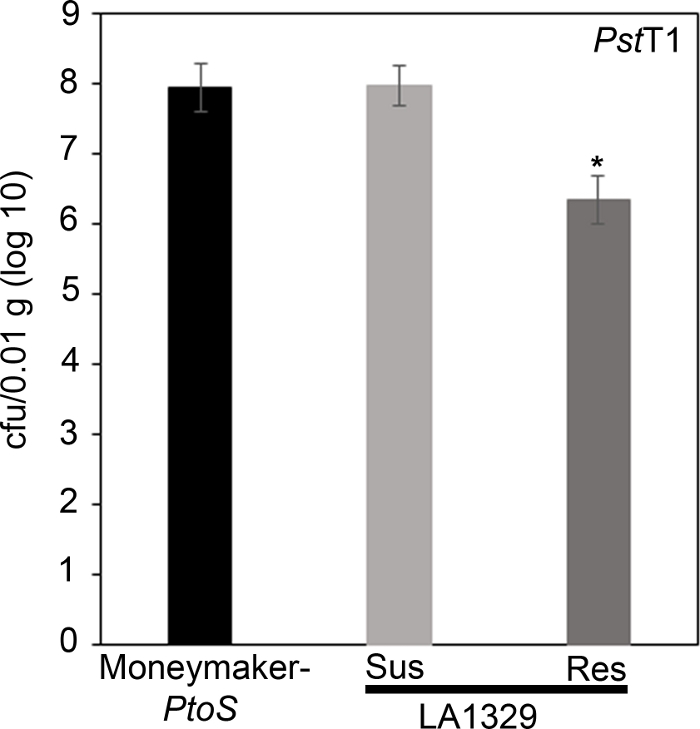
Figure 7: Resistant Solanum neorickii LA1329 seedlings support lower bacterial growth than Moneymaker-PtoS or susceptible S. neorickii LA1329. Bacterial counts were determined 4 days post-inoculation from S. neorickii LA1329 (n = 14) and Moneymaker-PtoS (n = 10) seedlings infected with PstT1 and normalization was performed to 0.01 g of tissue. For LA1329, the two phenotypic groups, susceptible (SUS) or resistant (RES), were observed and counted separately. Above the bar * = statistically significant difference determined by a one-factor analysis of variance. A general linear model procedure (p < 0.001) followed by a multiple comparison of means using Tukey's post hoc test was used. Error bars = standard error. The figure indicates one representative experiment. Please click here to view a larger version of this figure.
x
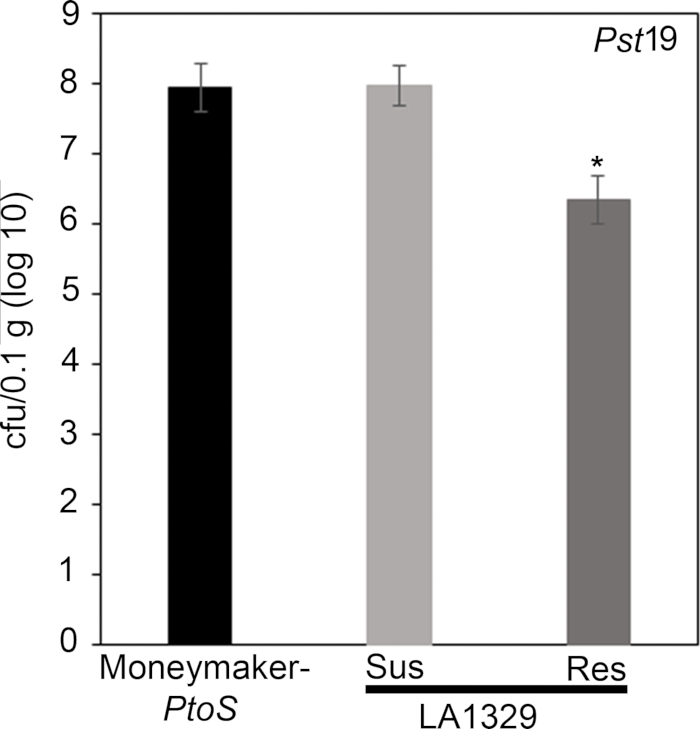
Figure 7: Resistant Solanum neorickii LA1329 seedlings support lower bacterial growth than Moneymaker-PtoS or susceptible S. neorickii LA1329. Bacterial counts were determined 4 days post-inoculation from S. neorickii LA1329 (n = 14) and Moneymaker-PtoS (n = 10) seedlings infected with Pst19 and normalization was performed to 0.1 g of tissue. For LA1329, the two phenotypic groups, susceptible (SUS) or resistant (RES), were observed and counted separately. Above the bar * = statistically significant difference determined by a one-factor analysis of variance. A general linear model procedure (p < 0.001) followed by a multiple comparison of means using Tukey's post hoc test was used. Error bars = standard error. The figure indicates one representative experiment. Please click here to view a larger version of this figure.
The first paragraph of the Discussion section was updated from:
A protocol for flood inoculation with PstDC3000 or PstT1 optimized to detect resistance to these bacterial strains in tomato seedlings is described. There are several critical parameters for optimal results in the seedling resistance assay, including bacterial concentration and surfactant concentration, which were empirically determined22. For PstDC3000, the optical density was optimized to achieve complete survival on a resistant cultivar containing the Pto/Prf cluster and complete death on a susceptible cultivar lacking the Pto/Prf cluster22. For a strain such as PstT1, where there are no known resistant varieties, the optical density was optimized to be the lowest possible for consistent and complete plant death22. Uppalapati et al.24 designed a tomato seedling assay to investigate the pathogenesis of PstDC3000 and the virulence function of coronatine. In this virulence assay, infections were performed using bacteria concentrated to an OD600 of 0.124, 20x higher than the optical density of strains used in our resistance assay. Recognition of PstDC3000 effectors AvrPto and AvrPtoB in tomato seedlings carrying the Pto/Prf gene cluster results in ETI and a macroscopic HR22. In the context of a strong immune response such as ETI, a lower bacterial titer was used for PstDC3000 to avoid overwhelming genetic resistance from the Pto/Prf gene cluster22. In addition, these results suggest that a high bacterial concentration could overwhelm weaker immune responses such as PTI or quantitative partial resistance, where multiple genes contribute to the overall phenotype. Surfactant is necessary for the bacteria to adhere to the leaf surface; however, high concentrations can cause chlorosis of the leaf22. We previously tested a range of surfactant concentrations to empirically determine the ideal concentration in 10-day-old tomato seedlings22. When testing new species that may differ in their sensitivity to surfactant, the surfactant concentration should be optimized to identify a concentration that does not cause damage or chlorosis in the absence of bacteria. Appropriate assay conditions will require optimization of a surfactant concentration that does not cause damage, and a bacterial concentration that causes disease in all susceptible controls.
to:
A protocol for flood inoculation with PstDC3000 or Pst19 optimized to detect resistance to these bacterial strains in tomato seedlings is described. There are several critical parameters for optimal results in the seedling resistance assay, including bacterial concentration and surfactant concentration, which were empirically determined22. For PstDC3000, the optical density was optimized to achieve complete survival on a resistant cultivar containing the Pto/Prf cluster and complete death on a susceptible cultivar lacking the Pto/Prf cluster22. For a strain such as Pst19, where there are no known resistant varieties, the optical density was optimized to be the lowest possible for consistent and complete plant death22. Uppalapati et al.24 designed a tomato seedling assay to investigate the pathogenesis of PstDC3000 and the virulence function of coronatine. In this virulence assay, infections were performed using bacteria concentrated to an OD600 of 0.124, 20x higher than the optical density of strains used in our resistance assay. Recognition of PstDC3000 effectors AvrPto and AvrPtoB in tomato seedlings carrying the Pto/Prf gene cluster results in ETI and a macroscopic HR22. In the context of a strong immune response such as ETI, a lower bacterial titer was used for PstDC3000 to avoid overwhelming genetic resistance from the Pto/Prf gene cluster22. In addition, these results suggest that a high bacterial concentration could overwhelm weaker immune responses such as PTI or quantitative partial resistance, where multiple genes contribute to the overall phenotype. Surfactant is necessary for the bacteria to adhere to the leaf surface; however, high concentrations can cause chlorosis of the leaf22. We previously tested a range of surfactant concentrations to empirically determine the ideal concentration in 10-day-old tomato seedlings22. When testing new species that may differ in their sensitivity to surfactant, the surfactant concentration should be optimized to identify a concentration that does not cause damage or chlorosis in the absence of bacteria. Appropriate assay conditions will require optimization of a surfactant concentration that does not cause damage, and a bacterial concentration that causes disease in all susceptible controls.
The third paragraph of the Discussion section was updated from:
Pst is a foliar pathogen that preferentially colonizes the aerial parts of tomato seedlings, including the cotyledons24 (Figure 3). Therefore, qualitative phenotyping in the seedling flood assay focuses on growth and disease symptoms in aerial portions of the seedling, and tissue for the bacterial growth assay is sampled from the cotyledons for quantitative analysis. After flood inoculation, seedlings may die within 7–10 days after inoculation with PstDC3000 or 10–14 days after inoculation with PstT1, as discussed in section 11. Seedling death is visualized by a brown apical meristem, arrested epicotyl elongation, and/or arrested vegetative growth. If different bacterial strains are used, the timing will have to be empirically determined. In addition, the progression of disease on control plants should be monitored daily after flooding until a consistent time frame from the onset of disease symptoms to seedling death can be identified. Depending on the genotypes and treatments used in the flood assay, seedling phenotypes can be recorded as binary phenotypes or on a disease spectrum (Figure 4). A broader spectrum of phenotypes may be observed when flood inoculating F2 mapping populations from wild tomato accessions crossed to susceptible cultivars (Figure 4C). It may be best to phenotype segregating populations on a disease spectrum depending on how quickly the seedling dies and the degree of new vegetative growth and branching (Figure 4C). The seedling flood assay can also be used in conjunction with the seedling bacterial growth assay to quantitatively assess levels of bacterial growth associated with qualitative phenotypes in individual seedlings (Figure 7). Very large reductions (i.e., ~log 3) in bacterial growth or strong resistance in resistant seedlings of a wild accession compared to a susceptible cultivar suggest that the underlying genetic basis of resistance may be due to ETI22. Smaller reductions in bacterial growth (i.e., ~log 1.7), as observed in LA1329 seedlings, may be due to the contribution of weaker resistance from quantitative trait loci and/or PTI. Thus, the seedling growth assay can be an important tool in further characterizing resistance in wild tomato lines.
to:
Pst is a foliar pathogen that preferentially colonizes the aerial parts of tomato seedlings, including the cotyledons24 (Figure 3). Therefore, qualitative phenotyping in the seedling flood assay focuses on growth and disease symptoms in aerial portions of the seedling, and tissue for the bacterial growth assay is sampled from the cotyledons for quantitative analysis. After flood inoculation, seedlings may die within 7–10 days after inoculation with PstDC3000 or 10–14 days after inoculation with Pst19, as discussed in section 11. Seedling death is visualized by a brown apical meristem, arrested epicotyl elongation, and/or arrested vegetative growth. If different bacterial strains are used, the timing will have to be empirically determined. In addition, the progression of disease on control plants should be monitored daily after flooding until a consistent time frame from the onset of disease symptoms to seedling death can be identified. Depending on the genotypes and treatments used in the flood assay, seedling phenotypes can be recorded as binary phenotypes or on a disease spectrum (Figure 4). A broader spectrum of phenotypes may be observed when flood inoculating F2 mapping populations from wild tomato accessions crossed to susceptible cultivars (Figure 4C). It may be best to phenotype segregating populations on a disease spectrum depending on how quickly the seedling dies and the degree of new vegetative growth and branching (Figure 4C). The seedling flood assay can also be used in conjunction with the seedling bacterial growth assay to quantitatively assess levels of bacterial growth associated with qualitative phenotypes in individual seedlings (Figure 7). Very large reductions (i.e., ~log 3) in bacterial growth or strong resistance in resistant seedlings of a wild accession compared to a susceptible cultivar suggest that the underlying genetic basis of resistance may be due to ETI22. Smaller reductions in bacterial growth (i.e., ~log 1.7), as observed in LA1329 seedlings, may be due to the contribution of weaker resistance from quantitative trait loci and/or PTI. Thus, the seedling growth assay can be an important tool in further characterizing resistance in wild tomato lines.
The fourth paragraph of the Discussion section was updated from:
Typically, genetic screens have been performed on four- to five-week-old adult tomato plants to identify the genetic basis of P. syringae resistance in wild accessions20,21. Adult tomato plants require much longer growth times, require more space in the growth chamber, and are much larger plants, which means that usually few individuals are screened for each line. The seedling flood assay provides a powerful, alternative approach in the identification of P. syringae resistance in wild tomato accessions. Screening at the seedling stage permits a large sample size to be tested which can be particularly advantageous in detecting resistance in genetically complex populations. Reduced growth chamber space requirements and growth time facilitate a high-throughput approach and rapid detection of natural resistance in wild accessions to emerging pathogens. Furthermore, P. syringae resistance that was identified at the seedling stage in this assay is not restricted to the developmental stage. S. neorickii LA1329 and S. habrochaites LA1253 were initially identified at the seedling stage and also display resistance to PstT1 in adult plants as previously described22.
to:
Typically, genetic screens have been performed on four- to five-week-old adult tomato plants to identify the genetic basis of P. syringae resistance in wild accessions20,21. Adult tomato plants require much longer growth times, require more space in the growth chamber, and are much larger plants, which means that usually few individuals are screened for each line. The seedling flood assay provides a powerful, alternative approach in the identification of P. syringae resistance in wild tomato accessions. Screening at the seedling stage permits a large sample size to be tested which can be particularly advantageous in detecting resistance in genetically complex populations. Reduced growth chamber space requirements and growth time facilitate a high-throughput approach and rapid detection of natural resistance in wild accessions to emerging pathogens. Furthermore, P. syringae resistance that was identified at the seedling stage in this assay is not restricted to the developmental stage. S. neorickii LA1329 and S. habrochaites LA1253 were initially identified at the seedling stage and also display resistance to Pst19 in adult plants as previously described22.
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。