Abbildung der Dehnungsbelastung eines Bauchaortenaneurysmas
Überblick
Quelle: Hannah L. Cebull1, Arvin H. Soepriatna1, John J. Boyle2 und Craig J. Goergen1
1 Weldon School of Biomedical Engineering, Purdue University, West Lafayette, Indiana
2 Mechanical Engineering & Materials Science, Washington University in St. Louis, St Louis, Missouri
Das mechanische Verhalten von Weichgeweben wie Blutgefäßen, Haut, Sehnen und anderen Organen wird stark durch ihre Zusammensetzung von Elastin und Kollagen beeinflusst, die Elastizität und Festigkeit bieten. Die Faserausrichtung dieser Proteine hängt von der Art des Weichgewebes ab und kann von einer einzigen bevorzugten Richtung bis hin zu komplizierten netzgebundenen Netzwerken reichen, die sich in krankem Gewebe verändern können. Daher verhalten sich Weichgewebe oft anisotrop auf Zell- und Organebene, was eine notwendigkeit dreidimensionale Charakterisierung erzeugt. Die Entwicklung einer Methode zur zuverlässigen Abschätzung von Dehnungsfeldern innerhalb komplexer biologischer Gewebe oder Strukturen ist wichtig, um Krankheiten mechanisch zu charakterisieren und zu verstehen. Dehnung stellt dar, wie Sichweich im Laufe der Zeit relativ verformt, und kann mathematisch durch verschiedene Schätzungen beschrieben werden.
Durch das Erfassen von Bilddaten im Laufe der Zeit können Verformungen und Dehnungen geschätzt werden. Alle medizinischen Bildgebungsmodalitäten enthalten jedoch eine gewisse Menge an Rauschen, was die Schwierigkeit erhöht, die in vivo-Stämme genau zu schätzen. Die hier beschriebene Technik bewindet diese Probleme erfolgreich, indem eine DDE-Methode (Direct Deformation Estimation) verwendet wird, um räumlich variierende 3D-Dehnungsfelder aus volumetrischen Bilddaten zu berechnen.
Zu den aktuellen Methoden zur Dehnungsschätzung gehören die digitale Bildkorrelation (DIC) und die digitale Volumenkorrelation. Leider kann DIC die Belastung von einer 2D-Ebene nur genau abschätzen, was die Anwendung dieser Methode stark einschränkt. Obwohl sie nützlich sind, haben 2D-Methoden wie DIC Schwierigkeiten, Dehnungen in Regionen zu quantifizieren, die einer 3D-Verformung unterzogen werden. Dies liegt daran, dass a-of-plane Bewegung Verformungsfehler erzeugt. Die digitale Volumenkorrelation ist eine anwendbarere Methode, die die anfänglichen Volumendaten in Regionen unterteilt und den ähnlichsten Bereich des verformten Volumens findet, wodurch Fehler a-of-plane reduziert werden. Diese Methode erweist sich jedoch als geräuschempfindlich und erfordert Annahmen über die mechanischen Eigenschaften des Materials.
Die hier gezeigte Technik beseitigt diese Probleme mit einer DDE-Methode und macht sie somit sehr nützlich bei der Analyse medizinischer Bildgebungsdaten. Darüber hinaus ist es robust bis hoch oder lokalisiert. Hier beschreiben wir die Erfassung von gated, volumetrischen 4D-Ultraschalldaten, deren Umwandlung in ein analyzierbares Format und die Verwendung eines benutzerdefinierten Matlab-Codes zur Abschätzung von 3D-Verformungen und entsprechenden Green-Lagrange-Stämmen, ein Parameter, der große Verformungen besser beschreibt. Der Green-Lagrange Dehnungstensor wird in vielen 3D-Dehnungsschätzungsmethoden implementiert, da er es ermöglicht, F aus einem Least Squares Fit (LSF) der Verschiebungen zu berechnen. Die folgende Gleichung stellt den Grün-Lagrange-Dehnungstensor Edar, wobei F und I den Verformungsgradienten bzw. den Identitätstensor zweiter Ordnung darstellen.
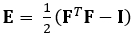 (1)
(1)
Verfahren
1. 4D-Ultraschall-Setup
- Verwenden Sie bei der Verwendung der Imaging-Software einen Laptop, der mathematische Computersoftware ausführt, um den 4D-Erfassungsprozess zu automatisieren. Schließen Sie den Laptop mit diesem benutzerdefinierten Code über den USB-Anschluss an das Ultraschallsystem an. Beachten Sie, dass die Bildverarbeitungssoftware über eine 4D-Ultraschallfunktion verfügt, die in die Software integriert ist.
- Nach dem Einschalten des Ultraschallsystems richten Sie die physiologische Überwac
Ergebnisse
Mit dem oben beschriebenen Verfahren wurde 4D-Ultraschall eines Angiotensin-II-induzierten suprarenalen sezierenden Abdominalaortenaneurysms (AAA) einer Maus erfasst. Entlang der Aorta wurden mehrere Kurzachs-EKV-Videoschleifen erfasst und zu 4D-Daten kombiniert, wie in Abbildung 1dargestellt. Diese Daten wurden dann mit einem benutzerdefinierten Code in eine MAT-Datei konvertiert, die dann in einem 3D-Stammberechnungscode mit einer Verzugsfunktion analysiert wurde. Nach der Optimierung der
Anwendung und Zusammenfassung
Die lokalisierte mechanische In-vivo-Charakterisierung ist ein wichtiger Bestandteil des Verständnisses des Wachstums und der Umgestaltung biologischer Gewebe. Im Vergleich zu bestehenden Ansätzen verwendet das hier beschriebene Dehnungsquantifizierungsverfahren eine verbesserte Methode zur genauen Berechnung der 3D-Dehnung durch optimale Verzug des unverformten Bildes vor der Kreuzkorrelation. Bei dieser Methode werden keine wesentlichen Annahmen zur Bestimmung von Stämmen innerhalb von Gewebevolumina verwen...
pringen zu...
Videos aus dieser Sammlung:

Now Playing
Abbildung der Dehnungsbelastung eines Bauchaortenaneurysmas
Biomedical Engineering
4.6K Ansichten

Bildgebung biologischer Proben mit optischer und konfokaler Mikroskopie
Biomedical Engineering
35.8K Ansichten

SEM-Bildgebung biologischer Proben
Biomedical Engineering
23.8K Ansichten

Biodistribution von Nano-Wirkstoffträgern: Anwendungen von SEM
Biomedical Engineering
9.3K Ansichten

Hochfrequenz-Ultraschall-Bildgebung der Bauchaorta
Biomedical Engineering
14.5K Ansichten

Photoakustische Tomographie zur Darstellung von Blut und Lipiden in der infrarenalen Aorta
Biomedical Engineering
5.7K Ansichten

Kardiale Magnetresonanztomographie
Biomedical Engineering
14.8K Ansichten

Numerische Strömungsmechaniksimulationen des Blutflusses in einem zerebralen Aneurysma
Biomedical Engineering
11.8K Ansichten

Nahinfrarot-Fluoreszenz-Bildgebung von Abdominalaortenaneurysmen
Biomedical Engineering
8.3K Ansichten

Nichtinvasive Blutdruckmesstechniken
Biomedical Engineering
11.9K Ansichten

Erfassung und Analyse eines EKG-Signals (Elektrokardiographie)
Biomedical Engineering
105.6K Ansichten

Zugfestigkeit resorbierbarer Biomaterialien
Biomedical Engineering
7.5K Ansichten

Mikro-CT-Bildgebung von Maus-Rückenmark
Biomedical Engineering
8.0K Ansichten

Visualisierung der Kniegelenksdegeneration nach nicht-invasiver Kreuzbandverletzung bei Ratten
Biomedical Engineering
8.2K Ansichten

Kombinierte SPECT- und CT-Bildgebung zur Visualisierung der Herzfunktionalität
Biomedical Engineering
11.0K Ansichten
Copyright © 2025 MyJoVE Corporation. Alle Rechte vorbehalten