Se requiere una suscripción a JoVE para ver este contenido. Inicie sesión o comience su prueba gratuita.
Method Article
Aplicación del análisis factorial multiómico no supervisado para descubrir patrones de variación y procesos moleculares relacionados con las enfermedades cardiovasculares
En este artículo
Resumen
Presentamos un flujo de trabajo flexible y extensible basado en Jupyter-lab para el análisis no supervisado de conjuntos de datos multiómicos complejos que combina diferentes pasos de preprocesamiento, estimación del modelo de análisis factorial multiómico y varios análisis posteriores.
Resumen
Los mecanismos de las enfermedades suelen ser complejos y se rigen por la interacción de varios procesos moleculares distintos. Los conjuntos de datos complejos y multidimensionales son un recurso valioso para generar más información sobre esos procesos, pero el análisis de dichos conjuntos de datos puede ser un desafío debido a la alta dimensionalidad que resulta, por ejemplo, de diferentes condiciones de enfermedades, puntos de tiempo y ómicas que capturan el proceso a diferentes resoluciones.
Aquí, mostramos un enfoque para analizar y explorar un conjunto de datos multiómicos tan complejo de manera no supervisada mediante la aplicación del análisis factorial multiómico (MOFA) a un conjunto de datos generado a partir de muestras de sangre que capturan la respuesta inmune en síndromes coronarios agudos y crónicos. El conjunto de datos consta de varios ensayos con diferentes resoluciones, incluidos datos de citocinas a nivel de muestra, proteómica de plasma y secuenciación principal de neutrófilos, y datos de secuenciación de ARN de una sola célula (scRNA-seq). Se añade más complejidad al tener varios puntos de tiempo diferentes medidos por paciente y varios subgrupos de pacientes.
El flujo de trabajo de análisis describe cómo integrar y analizar los datos en varios pasos: (1) Preprocesamiento y armonización de datos, (2) Estimación del modelo MOFA, (3) Análisis posterior. En el paso 1 se describe cómo procesar las características de los diferentes tipos de datos, filtrar las características de baja calidad y normalizarlas para armonizar sus distribuciones para un análisis posterior. El paso 2 muestra cómo aplicar el modelo MOFA y explorar las principales fuentes de varianza dentro del conjunto de datos en todas las ómicas y características. En el paso 3 se presentan varias estrategias para el análisis posterior de los patrones capturados, vinculándolos a las condiciones de la enfermedad y a los posibles procesos moleculares que gobiernan esas condiciones.
En general, presentamos un flujo de trabajo para la exploración de datos no supervisada de conjuntos de datos multiómicos complejos para permitir la identificación de los principales ejes de variación compuestos por diferentes características moleculares que también se pueden aplicar a otros contextos y conjuntos de datos multiómicos (incluidos otros ensayos presentados en el caso de uso ejemplar).
Introducción
Los mecanismos de las enfermedades suelen ser complejos y se rigen por la interacción de varios procesos moleculares distintos. Descifrar los complejos mecanismos moleculares que conducen a enfermedades específicas o gobiernan la evolución de una enfermedad es una tarea de gran relevancia médica, ya que podría revelar nuevos conocimientos para la comprensión y el tratamiento de las enfermedades.
Los avances tecnológicos recientes permiten medir esos procesos en una resolución más alta (por ejemplo, a nivel de una sola célula) y en varias capas biológicas (por ejemplo, ADN, ARNm, accesibilidad a la cromatina, metilación del ADN, proteómica) al mismo tiempo. Esto conduce a la generación cada vez mayor de grandes conjuntos de datos biológicos multidimensionales, que se pueden analizar conjuntamente para generar más información sobre los procesos subyacentes. Al mismo tiempo, combinar y analizar las diferentes fuentes de datos de una manera biológicamente significativa sigue siendo una tarea desafiante1.
Los diferentes límites tecnológicos, ruidos y rangos de variabilidad entre las diferentes ómicas plantean un desafío. Por ejemplo, los datos de secuenciación de ARN de una sola célula (scRNA-seq) son muy escasos y, a menudo, están influenciados por grandes efectos técnicos o de lotes. Además, el espacio de características suele ser muy grande, abarcando varios miles de genes o proteínas medidos, mientras que los tamaños de muestra son limitados. Esto se complica aún más por los diseños complejos, que pueden incluir varias condiciones de enfermedad, factores de confusión, puntos de tiempo y resoluciones. Por ejemplo, en el caso de uso presentado, había diferentes tipos de datos disponibles a nivel de una sola célula o de muestra (a granel). Además de eso, los datos pueden estar incompletos y es posible que no todas las mediciones estén disponibles para todos los sujetos analizados.
Debido a estos desafíos, las diferentes ómicas y las características incluidas a menudo se analizan solo por separado2, aunque la realización de un análisis integrado no solo puede proporcionar una imagen completa del proceso, sino que los ruidos biológicos y técnicos de una ómica también pueden ser compensados por otras ómicas 3,4. Se han propuesto varios métodos diferentes para realizar un análisis integrado de datos multiómicos, incluidos los métodos bayesianos, los métodos basados en redes 5,6, el aprendizaje profundo multimodal7 y los métodos de reducción de dimensionalidad mediante factorización de matrices 8,9. Para este último, los resultados de un gran estudio de benchmarking10 han demostrado que el método MOFA9 (análisis factorial multiómico) es una de las herramientas más adecuadas cuando los datos deben vincularse a las anotaciones clínicas.
Especialmente en entornos complejos, los métodos de factorización de matrices no supervisados son un enfoque útil para reducir la complejidad y extraer señales compartidas y complementarias de diferentes fuentes de datos y características. Al descomponer el espacio complejo en representaciones latentes de rango inferior, las principales fuentes de varianza dentro de los datos se pueden explorar rápidamente y vincular a covariables conocidas. En caso de que el mismo patrón de variación se comparta a través de múltiples características (por ejemplo, genes o proteínas), esto puede agregarse a unos pocos factores mientras se reduce el ruido. La regularización se puede utilizar para aumentar la dispersión de los coeficientes del modelo, lo que hace que el enfoque sea adecuado en entornos en los que el espacio de características es grande mientras que el número de muestras es limitado9.
Este protocolo presenta un flujo de trabajo de análisis flexible que utiliza el modelo MOFA para mostrar cómo explorar rápidamente un conjunto de datos multiómicos complejo y destilar los principales patrones de variación que caracterizan a este conjunto de datos. El flujo de trabajo consta de tres pasos principales. En el primer paso, Preprocesamiento y armonización de datos, se presentan diferentes estrategias para el preprocesamiento de datos basadas en diferentes tipos de datos de entrada (scRNA-seq, proteómica, citocinas, datos clínicos). El protocolo explica cómo procesar las características de los diferentes conjuntos de datos de entrada, filtrar las características de baja calidad y normalizarlas para armonizar sus distribuciones. También mostramos cómo esas decisiones de preprocesamiento pueden afectar a los resultados posteriores. En el segundo paso, se aplica el modelo MOFA a los datos, y la descomposición de la varianza resultante se puede utilizar para evaluar la integración de los diferentes conjuntos de datos. El tercer paso muestra cómo vincular los factores capturados con las covariables y descubrir los programas moleculares que definen esos factores. Con el flujo de trabajo presentado, pudimos extraer varios factores latentes vinculados a covariables clínicas en un conjunto de datos de pacientes que padecían síndromes coronarios e identificar posibles programas inmunes multicelulares subyacentes de un proyecto anterior11. Usaremos este conjunto de datos aquí, pero el protocolo se puede aplicar fácilmente a otros contextos, incluidas otras ómicas.
El conjunto de datos consta de muestras de pacientes con síndromes coronarios crónicos (SCC) estables, síndromes coronarios agudos (SCA) y un grupo de control con coronarias sanas (no SCC) (Figura 1). El SCA es causado por la ruptura de la placa en el CCS preexistente, lo que conduce a una interrupción aguda del flujo sanguíneo al miocardio y una lesión isquémica posterior del corazón. Esta lesión provoca una respuesta inflamatoria del sistema inmune seguida de una fase reparadora, que dura hasta varios días después del evento agudo12. Para poder caracterizar esta respuesta inmunitaria en los pacientes con SCA, se tomaron muestras de sangre en cuatro momentos diferentes: agudo (TP1); después de la recanalización (14 [± 8] h) (TP2); 60 [± 12] h después (TP3); antes del alta (6,5 [±1,5] días) (TP4) (Figura 1A). Para el CCS y los pacientes con coronarias sanas, solo se disponía de un punto de tiempo: (TP0). Para todos los pacientes y puntos temporales se midieron diferentes ensayos basados en las muestras de sangre: marcadores clínicos de inflamación (creatina-quinasa (CK), CK-MB, TROPONINA, PROTEÍNA C reactiva (PCR)), scRNA-seq de células mononucleares de sangre periférica (PBMC), análisis de citocinas, proteómica plasmática y datos prime-seq13 de neutrófilos.
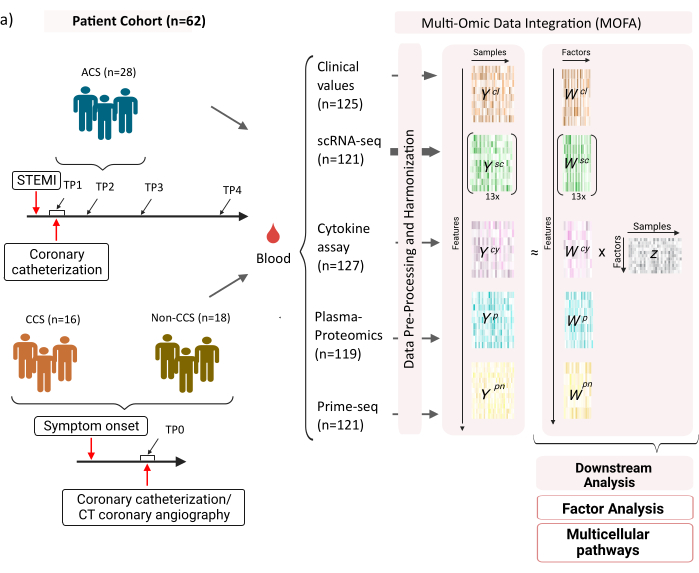
Figura 1: Conjunto de datos de entrada multiómica de infarto de miocardio. Conjunto de datos de entrada: Los datos analizados incluyen muestras de sangre de pacientes (n = 62) con síndrome coronario agudo (SCA), síndromes coronarios crónicos (SCC) y pacientes con coronarias sanas (no SCC). Para los pacientes con SCA, se incluyeron muestras de sangre en cuatro puntos de tiempo diferentes (TP1-4), para los pacientes con CCS y no CCS en un solo punto de tiempo (TP0). Cada combinación de paciente y punto de tiempo se trata como una muestra separada en el análisis. Se midieron diferentes ensayos ómicos en las muestras: análisis clínicos de sangre (n = 125), scRNA-seq (n = 121), plasma-proteómica (n = 119), ensayo de citocinas (n = 127) y neutrophil prime-seq (n = 121). Posteriormente, se aplicó el protocolo descrito para integrar los datos en todas las ómicas y explorarlos utilizando el modelo MOFA y análisis posteriores (análisis factorial, enriquecimiento de vías). Haga clic aquí para ver una versión más grande de esta figura.
Como entrada para el flujo de trabajo que se presenta aquí, tomamos recuentos brutos de los datos de scRNA-seq después del procesamiento con cellranger y el control de calidad (QC) como, por ejemplo, se describe en el tutorial de preprocesamiento de scanpy14 . Para la anotación del tipo de celda, utilizamos la canalización automatizada Azimut15 . A continuación, los recuentos se agregan a nivel de muestra para cada tipo de célula tomando la media de todas las células de cada muestra y tipo de célula (agregación pseudomasiva). La proteómica del plasma se incluye como intensidades normalizadas y centradas en la mediana, y para los neutrófilos, tomamos los recuentos de exones del identificador molecular único (UMI) de la secuencia principal. Sobre citocinas y valores clínicos, no se ha aplicado ningún preprocesamiento previo. En el manuscrito correspondiente11 se describen más detalles sobre la generación de datos (experimentales). Dado que los resultados presentados aquí se basan en el uso de la anotación de acimut automatizada para los tipos de células en los datos de scRNA-seq en comparación con la estrategia basada en marcadores que se utilizó en la publicación a la que se hace referencia, los resultados presentados aquí son similares, pero no exactamente iguales a los presentados en la publicación. En el manuscrito se pudo demostrar que la estrategia de anotación del tipo de celda no cambia los patrones principales y las interpretaciones biológicas del análisis, pero pequeños cambios en los valores exactos resultantes del modelo pueden variar. En general, los datos de entrada eran un conjunto de datos complejo y multidimensional que incluía diferentes puntos de tiempo y niveles de medición (células individuales frente a graneles) de más de 10.000 características diferentes (genes, proteínas, valores clínicos). Se ha demostrado que una estricta estrategia de preprocesamiento y armonización de datos seguida por el análisis MOFA es una herramienta útil y rápida para explorar los datos y extraer el programa inmunológico relevante. Cada combinación de punto de tiempo y paciente se trata como una muestra independiente en el análisis MOFA. Cada tipo de datos y tipo de celda se considera una vista separada en el análisis MOFA.
Este protocolo proporciona instrucciones para preparar los datos de entrada para el flujo de trabajo, ejecutar los diferentes pasos del flujo de trabajo, personalizar las configuraciones, interpretar las cifras resultantes y ajustar iterativamente las configuraciones en función de las interpretaciones. En la descripción general del flujo de trabajo técnico (Figura 2) se proporciona una descripción general de los diferentes pasos del protocolo, los conjuntos de datos de entrada necesarios en cada paso y las figuras y conjuntos de datos resultantes.
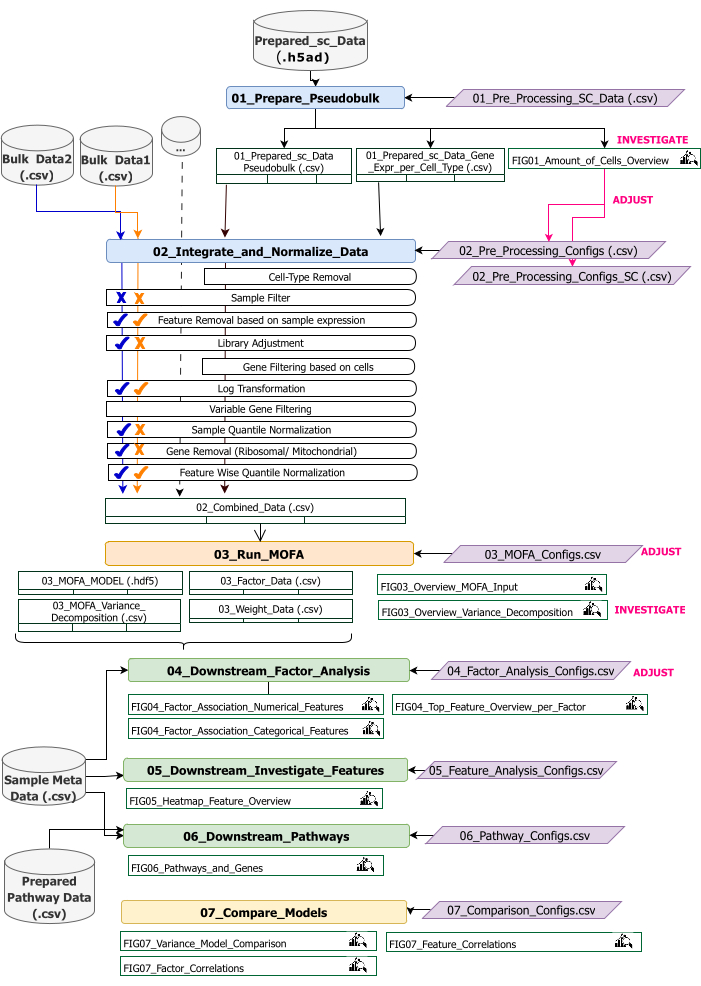
Figura 2: Descripción general del flujo de trabajo técnico. Esquema del flujo de trabajo para el análisis del conjunto de datos multiómicos. Los diferentes elementos están resaltados por diferentes colores y símbolos. Los Jupyter Notebooks que pertenecen al paso Preprocesamiento y armonización de datos (1) están coloreados en azul. Los Jupyter Notebooks que pertenecen al paso 'Modelo MOFA' (2) están coloreados en naranja. Los Jupyter Notebooks que pertenecen al paso "Análisis descendente" (3) están coloreados en verde. Un cuaderno de Jupyter Notebook que se usará para comparar los resultados está coloreado en amarillo. Los archivos de configuración donde se pueden modificar los parámetros para la ejecución del flujo de trabajo se resaltan en púrpura. Los conjuntos de datos de entrada necesarios para ejecutar el flujo de trabajo se indican mediante el símbolo del conjunto de datos y se resaltan en gris. Todas las salidas de figuras que se generan durante la ejecución del flujo de trabajo se indican con el símbolo de lupa. Los conjuntos de datos generados durante la ejecución del flujo de trabajo se indican como tablas. En general, el flujo de trabajo se ejecuta secuencialmente: (1) El preprocesamiento y la armonización de datos consta de dos pasos: la primera generación de una tabla pseudomasiva basada en los datos de entrada scRNA-seq (01_Prepare_Pseudobulk) y la posterior integración y normalización de estos datos junto con todas las demás entradas de nivel de muestra (masivas) (02_Integrate_and_Normalize_Data). Dentro de este paso, a través de los archivos de configuración, es posible configurar para cada conjunto de datos por separado cuál de los pasos de preprocesamiento y normalización indicados (por ejemplo, filtro de muestra) se debe aplicar. (2) 'Modelo MOFA': ejecuta el modelo MOFA en la entrada generada del primer paso con las configuraciones especificadas en el archivo de configuración (03_MOFA_configs.csv) (3) 'Análisis descendente': consta de tres cuadernos diferentes que pueden ejecutarse independientemente entre sí para generar información sobre los resultados MOFA generados y asociarlos con metadatos de muestra (covariables) proporcionados como entrada a través del archivo 'Sample Meta Data.csv'. (4) "Comparación de modelos": es un pequeño paso separado que se puede utilizar para comparar diferentes modelos generados en el paso 2. Haga clic aquí para ver una versión más grande de esta figura.
El flujo de trabajo consta de varios Jupyter Notebooks escritos en R y Python (no se requiere conocimiento del lenguaje R y Python para ejecutar el flujo de trabajo, pero puede ser útil en caso de que aparezcan errores). En varios pasos del protocolo, los parámetros se modifican a través de archivos de configuración (archivos '.csv' que contienen el sufijo '_Configs' en el nombre). Dentro del protocolo, solo describimos los parámetros que deben cambiarse a partir de la configuración predeterminada.
También se pueden cambiar otros parámetros, por ejemplo, para personalizar el preprocesamiento. Una documentación de estos parámetros y explicaciones se proporciona en el archivo 'Documentation_Config_Parameter', que se incluye en el repositorio descargado.
Protocolo
1. Preparativos: Configuración técnica e instalación
NOTA: Para ejecutar este programa, tenga wget, git y Apptainer preinstalados en el dispositivo. Aquí se proporciona una guía para instalar Apptainer en diferentes sistemas (Linux, Windows, Mac): https://apptainer.org/docs/admin/main/installation.html. La información de instalación de git se puede encontrar aquí: https://git-scm.com/book/en/v2/Getting-Started-Installing-Git. En función del tamaño de los diferentes conjuntos de datos de entrada, se recomienda ejecutar el flujo de trabajo en un equipo adecuado (16 CPU, 64 GB de memoria). Una prueba de humo con los datos de ejemplo proporcionados se puede ejecutar en la máquina local. Las instrucciones y los resultados esperados de la ejecución del protocolo en los datos de ejemplo se proporcionan en el Archivo Suplementario 1. Consulte el Archivo de video complementario 1 para conocer los pasos importantes del protocolo que se ejecutan en el conjunto de datos descrito anteriormente.
- Abra la consola y elija o cree una carpeta donde se almacenarán todo el código de análisis y las salidas. Navegue a la carpeta escribiendo el comando: cd path_to_folder en el terminal.
- Descargue o clone el repositorio de código de Github (https://github.com/heiniglab/mofa_workflow) o escribiendo git clone https://github.com/heiniglab/mofa_workflow.git en la ventana del terminal.
- Descargue la imagen que contiene todas las instalaciones necesarias de Zenodo escribiendo wget https://zenodo.org/records/11192947/files/mofa_image.sif en la ventana del terminal.
- Genere una carpeta en la que se almacenarán todos los datos de los resultados escribiendo mkdir results en la ventana del terminal.
- Genere una carpeta en la que se agregarán todos los datos de entrada que se utilizarán en el análisis escribiendo mkdir input_data en la ventana del terminal.
- Ejecute el contenedor que iniciará una sesión de JupyterLab escribiendo el siguiente comando en el terminal: apptainer run mofa_image.sif. Copie la URL devuelta por el comando en el navegador, lo que abrirá una sesión de Jupyter-lab (puede encontrar más información sobre Jupyter-lab en la documentación del software16).
NOTA: Cuando el flujo de trabajo se ejecuta localmente en una computadora portátil, se recomienda utilizar el comando apptainer exec mofa_image.sif jupyter-lab en su lugar, que devolverá directamente una dirección de host local. En caso de que el contenedor se ejecute dentro de un entorno de computación en clúster, puede ser necesario configurar el reenvío de puertos, lo que se puede hacer a través de ssh.
2. Inicialización y preparación de datos
- En la sesión de Jupyter-Lab, use el menú de navegación del lado izquierdo. Navegue a la carpeta input_data haciendo doble clic en input_data.
- Copie todos los conjuntos de datos que se utilizarán como entrada para el análisis en el directorio input_data mediante la función de arrastrar y soltar. Arrastre el archivo desde la carpeta en la que se encuentra actualmente y suéltelo en la sesión de Jupyter-lab en el área debajo de la carpeta input_data.
NOTA: Todos los conjuntos de datos deben estar en formato .csv o .h5ad (en el caso de datos de una sola celda). Todos los archivos .csv deben contener una columna sample_id coincidente (se deben usar identificadores idénticos en todos los conjuntos de datos). Todas las demás columnas se usarán como características. Dentro del archivo h5ad-, la anotación de celda debe contener dos identificadores que especifiquen el sample_id y el cluster_id. Estos se utilizarán para la agregación y la coincidencia. Los conjuntos de datos ómicos en otros formatos deben convertirse al formato .csv especificado antes de su uso (Figura 3). Los conjuntos de datos scRNA-seq proporcionados en formato .h5seurat podrían convertirse a .h5ad ejecutando el Jupyter-notebook: 00_Data_Conversion.ipynb. - Navegue a la carpeta de configuraciones haciendo clic en el símbolo de la carpeta y, a continuación, haga doble clic en las carpetas mofa_workflow, scripts y configuraciones. Dentro de la carpeta, abra el archivo Data_configs.csv haciendo doble clic en él.
- En la columna de valor , agregue las rutas de acceso a las carpetas de las carpetas input_data (data_path) y resultados (result_path). Agregue un nombre que se agregará como una extensión de archivo a todos los archivos guardados en la columna de valor para el configuration_name (este protocolo se utilizó MI_v1 [Infarto de miocardio versión 1]) (Figura 4).
- Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV en el menú de la parte superior.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de scripts haciendo clic en scripts. Abra el cuaderno de inicialización haciendo doble clic en 00_Configuration_Update.ipynb. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior y haciendo clic en Reiniciar en la ventana emergente (Figura 5).
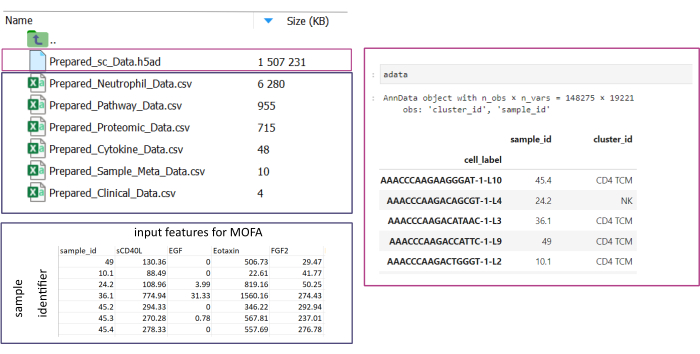
Figura 3: Entrada de datos y configuración. Para la ejecución del flujo de trabajo, todos los datos deben almacenarse en una carpeta input_data especificada. Para cada conjunto de datos de entrada, se debe proporcionar un archivo independiente. Los datos de una sola celda deben proporcionarse como .h5ad que contenga una anotación de celda en el cluster_id (resultante, por ejemplo, de pasos anteriores de anotación de tipo de celda) y una columna de sample_id (que identifique de manera única cada muestra separada que debe analizarse). Todos los demás conjuntos de datos de entrada deben proporcionarse en formato «.csv», incluida una columna que especifique la sample_id (que coincida con la columna correspondiente de los datos de una sola celda) y las características que se utilizarán en el análisis del MOFA en todas las demás columnas. Haga clic aquí para ver una versión más grande de esta figura.
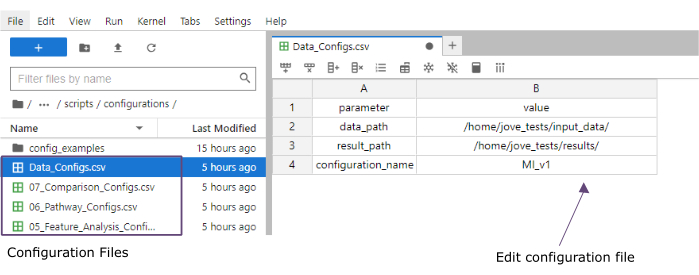
Figura 4: Archivos de configuración de Jupyter-lab. Durante la ejecución del flujo de trabajo, los cambios en los parámetros (por ejemplo, ajuste de las opciones de filtrado, etc.) se especifican a través de archivos de configuración '.csv'. Dentro del repositorio clonado, se incluyen los archivos de configuración predeterminados para cada paso. Se pueden editar directamente en la consola de jupyter-lab, de forma similar a una hoja de cálculo. Haga clic aquí para ver una versión más grande de esta figura.
Figura 5: Scripts de Jupyter-notebooks. El flujo de trabajo completo consta de una serie de cuadernos de Jupyter Notebook que se ejecutarán secuencialmente después de que se hayan modificado los archivos de configuración correspondientes. Al hacer doble clic en el cuaderno de Jupyter Notebook en el lado izquierdo, se abrirá el archivo correspondiente en el lado derecho. La ejecución completa del archivo se puede iniciar con el botón resaltado en la parte superior. Haga clic aquí para ver una versión más grande de esta figura.
3. Pretratamiento y armonización de datos
- Preprocesamiento: convierta los datos sc en pseudobulk.
NOTA: Este paso solo debe ejecutarse si se utilizan datos de una sola celda en el análisis.- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de configuraciones haciendo doble clic en configuración. Abra el archivo 01_Preprocessing_SC_Data.csv haciendo doble clic. Compruebe los valores rellenados automáticamente en el archivo y, si es necesario, ajuste los valores de la columna data_name para que se correspondan con los nombres de archivo de los conjuntos de datos de una sola celda en la carpeta input_data que se utilizará para el análisis.
NOTA: De forma predeterminada, todos los nombres de los archivos .h5ad en la carpeta de datos de entrada se agregarán al archivo de configuración en el script de inicialización. Si algunos de los conjuntos de datos no se deben utilizar para el análisis, se pueden eliminar aquí. - Guarde los cambios realizados haciendo clic en Archivo > Guardar archivo CSV en el menú de la parte superior.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de scripts haciendo clic en scripts. Abra el bloc de notas 01_Prepare_Pseudobulk.ipynb haciendo doble clic en él. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior y haciendo clic en Reiniciar en la ventana emergente.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de figuras haciendo doble clic primero en figuras y luego en 01_figures. Abra el FIG01_Amount_of_Cells_overview de gráfico recién generado haciendo doble clic en él.
NOTA: La ejecución del bloc de notas puede tardar varios minutos. Cuando el bloc de notas se haya ejecutado correctamente, aparecerá una ventana emergente y el archivo FIG01_Amount_of_Cells_Overview será actualizado por el bloc de notas o generado de nuevo. La columna Última modificación puede indicar cuándo se generó el archivo para evaluar si se trata de un archivo nuevo o antiguo. - Investigue la gráfica para identificar grupos de tipos de células con un número muy bajo de células por muestra. Anote los nombres de esos cluster_ids excluirlos en los pasos siguientes (Figura 6).
- Utilice el menú de navegación del lado izquierdo para volver a la carpeta de configuraciones haciendo clic en ... y luego haciendo doble clic en configuraciones. Abra el archivo 02_Preprocessing_Configs_SC.csv haciendo doble clic en él.
- Compruebe los valores de las columnas configuration_name y data_name y ajústelos si es necesario.
NOTA: Dentro del script de inicialización, esos valores se rellenan previamente con todos los nombres de los archivos .h5ad en la carpeta de datos de entrada y el valor de configuration_name establecido anteriormente en el archivo Data_Configs.csv . En caso de que los archivos deban excluirse del análisis o se deba utilizar otra extensión para los nombres de archivo, esto se puede ajustar aquí. - Ajuste el valor de la columna cell_type_exclusion y agregue todos los cluster_id que se hayan identificado para excluir en el paso anterior separados por ','.
- Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV en la barra de navegación de la parte superior.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de configuraciones haciendo doble clic en configuración. Abra el archivo 01_Preprocessing_SC_Data.csv haciendo doble clic. Compruebe los valores rellenados automáticamente en el archivo y, si es necesario, ajuste los valores de la columna data_name para que se correspondan con los nombres de archivo de los conjuntos de datos de una sola celda en la carpeta input_data que se utilizará para el análisis.
- Preprocesamiento: armonice e integre otras fuentes de datos ómicos.
- Abra el archivo 02_Preprocessing_Configs.csv haciendo doble clic sobre él y ajuste la configuración de preprocesamiento para cada uno de los conjuntos de datos que se incluirán y se almacena en la carpeta data_input (una fila por conjunto de datos).
- Compruebe los valores de las columnas configuration_name y data_name y ajústelos si es necesario.
- Ajuste los demás parámetros de las columnas en consecuencia, en función de los pasos de preprocesamiento que se deban aplicar.
NOTA: Los valores predeterminados se agregan para cada conjunto de datos que se encuentra dentro de la carpeta input_dataset, pero no son específicos de los tipos de datos individuales de los datos. Por lo tanto, serán necesarios ajustes. En el archivo de Documentation_Config_Parameter.doc se proporciona una documentación detallada de los parámetros. - Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de scripts haciendo clic en scripts. Abra el cuaderno 02_Integrate_and_Normalized_Data_Sources.ipynb haciendo doble clic en él. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior y haciendo clic en Reiniciar en la ventana emergente.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de 02_results generada haciendo clic en el símbolo de la carpeta y luego haciendo doble clic en resultados y 02_results. Compruebe si incluye el archivo 02_Combined_data_'configuration_name'_Integrated.csv que contiene el archivo de entrada de datos preprocesado combinado.
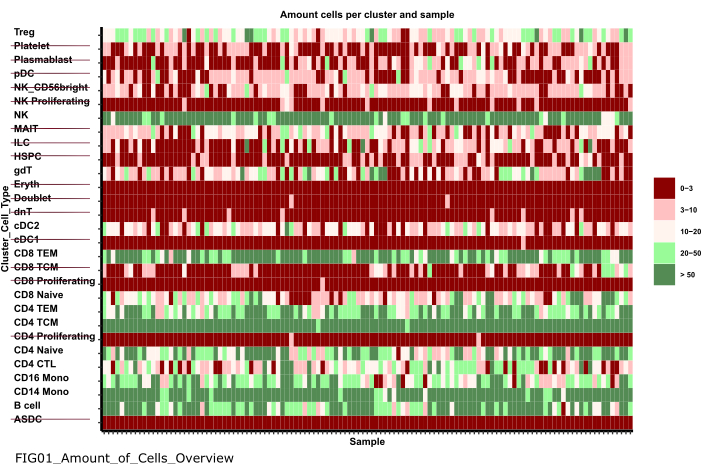
Figura 6: Preprocesamiento y armonización de datos. Una salida del paso '01_Prepare_Pseudobulk' es el gráfico 'Fig01_Amount_of_Cells_Overview'. Aquí, para cada cluster_id (eje Y que indica el tipo de célula de los pasos anteriores de anotación del tipo de célula), se da el número de células por muestra ('sample_id'). Dentro de los resultados presentados, los tipos de células con una baja cantidad de células por muestra se excluyen del análisis posterior (indicado por el tachado). Haga clic aquí para ver una versión más grande de esta figura.
4. Ejecutar MOFA
- En Jupyter-Lab, utilice el menú de navegación de la izquierda para navegar a la carpeta de configuraciones haciendo clic en el símbolo de la carpeta y, a continuación, haga doble clic en mofa_workflow, seguido de un doble clic en scripts y configuraciones. Abra el archivo 03_MOFA_Configs.csv haciendo doble clic en él.
- Compruebe las entradas de las columnas configuration_name y mofa_result_name y ajuste las entradas si se deben utilizar nombres alternativos.
NOTA: La mofa_result_name se agregará como una extensión de archivo a todos los archivos de resultados generados en función del MOFA. Esto puede ser diferente del valor configuration_name , ya que se pueden ejecutar diferentes configuraciones de MOFA con los mismos datos de entrada (este protocolo utiliza MI_v1_MOFA). - Introduzca la cantidad de factores que se deben estimar en el modelo MOFA (columna amount_of_factors ) y defina si se deben aplicar la ponderación y la escala (columnas weighting_of_views y scale_views ) ajustando los valores del archivo.
- Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de navegación en el lado izquierdo para navegar a la carpeta de scripts haciendo clic en 'scripts'. Abra el cuaderno 03_Run_MOFA.ipynb haciendo doble clic en el archivo. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior y haciendo clic en Reiniciar en la ventana emergente.
- Navegue a la carpeta 03_figures haciendo doble clic en las figuras y luego 03_figures. Abra el FIG03_Overview_Variance_Decomposition_mofa_result_name de la gráfica generada e investigue el resultado del modelo (Figura 7A).
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de 03_results generada haciendo clic en el símbolo de la carpeta y luego haciendo doble clic en resultados y 03_results. Compruebe si incluye el archivo de valor de factor de muestra 03_Factor_Data_'mofa_result_name'.csv y el archivo de peso de factor de característica 03_Weight_Data_'mofa_result_name'.csv.
5. Análisis posterior
- Interpretación factorial.
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta input_data haciendo clic en el símbolo de la carpeta , seguido de un doble clic en input_data.
- Preparar un fichero .csv (Prepared_Sample_Meta_Data.csv) que contenga todos los metadatos (covariables) de las muestras que se analizarán en asociación con los factores generados. Copie el archivo en la carpeta input_data mediante arrastrar y soltar el archivo en la vista general de la carpeta input_data .
NOTA: Debe contener la columna sample_id para que coincida con los datos utilizados anteriormente y columnas adicionales para cada característica que se debe analizar. - En Jupyter-Lab, utilice el menú de navegación de la izquierda para volver a la carpeta de configuraciones haciendo clic en el símbolo de la carpeta y, a continuación, haciendo doble clic en mofa_workflow, seguido de scripts y configuración. Abra el archivo 04_Factor_Analysis.csv haciendo doble clic en él.
- Compruebe que las entradas para el configuration_name y el mofa_result_name contengan los nombres de la configuración y los resultados de MOFA que se analizarán en el script y ajústelos si es necesario.
- En la columna numeric_covariates , agregue el nombre de todas las columnas numéricas del archivo Prepared_Sample_Meta_Data.csv que se investigarán en relación con los factores MOFA separados por comas (este protocolo utiliza CRP,CK).
- En la columna categorical_covariates', agregue el nombre de todas las columnas categóricas del archivo Prepared_Sample_Meta_Data.csv que se investigarán en relación con los factores MOFA separados por comas (este protocolo utiliza medición).
- Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de navegación de la izquierda para navegar a la carpeta 'scripts' haciendo clic en scripts. Abra el cuaderno 04_Downstream_Factor_Analysis.ipynb haciendo doble clic en él. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior y haciendo clic en reiniciar en la ventana emergente.
- Utilice el menú de navegación de la izquierda para navegar a la carpeta 04_figures haciendo doble clic en las figuras y luego en 04_figures. Abra los gráficos generados haciendo doble clic en ellos e investigue los factores para patrones y asociaciones interesantes: FIG04_Factor_Association_with_numeric_features_
'mofa_result_name.pdf (Figura 7B). FIG04_Factor_Association_
with_categorical_features_ mofa_result_name.pdf (Figura 7C). FIG04_Top_Feature_Overview_per_Factor _'mofa_result_name.pdf (Figura 8A).
- Análisis de características
- Utilice el menú de navegación de la izquierda para volver a la carpeta de configuraciones haciendo clic en ... y luego haciendo doble clic en configuraciones. Abra el archivo 05_Feature_Analysis_Configs.csv haciendo doble clic en él.
- Compruebe que las entradas de las columnas configuration_name y mofa_result_name correspondan a los nombres de la configuración y el resultado MOFA generado que se utilizará para el análisis posterior y ajústelas si es necesario.
- En la columna de factores , agregue el factor para el que se trazarán las características principales en el siguiente script.
- En la faceting_variable de columnas, agregue un nombre de columna de una columna categórica en el Prepared_Sample_Meta_Data.csv que se utilizará para agrupar las muestras en la gráfica (este protocolo utiliza la medición)
- Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de navegación de la izquierda para navegar a la carpeta de scripts haciendo clic en scripts. Abra el cuaderno 05_Downstream_Investigate_Features_Heatmap.ipynb haciendo doble clic en él. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior y haciendo clic en Reiniciar en la ventana emergente.
- Utilice el menú de navegación de la izquierda para navegar a la carpeta 05_figures haciendo doble clic primero en las figuras y luego en 05_figures. Abra e investigue la trama generada FIG05_Heatmap_Feature_Overview__ 'mofa_result_name'.pdf haciendo doble clic en el archivo (Figura 8B).
NOTA: Dependiendo de la cantidad de características que se mostrarán en el gráfico, puede ser necesario ajustar los parámetros plot_width y plot_height dentro del 05_Feature_Analysis_Configs.csv y volver a ejecutar el script para asegurarse de que todo encaja en el gráfico.
- Análisis de vías
- Utilice el menú de navegación del lado izquierdo para navegar a la carpeta input_data haciendo clic en el símbolo de la carpeta , seguido de un doble clic en input_data.
- Prepare un archivo de .csv (Prepared_Pathway_Data.csv) que contenga una lista de rutas de acceso que se probarán para el enriquecimiento. Copie el archivo en la carpeta input_data mediante arrastrar y soltar el archivo en la vista general de la carpeta input_data.
NOTA: Debe contener tres columnas: ID (un identificador único de la vía), gen (los genes dados por su nombre de gen (SÍMBOLO) que pertenecen a la vía, una fila por gen), pathway_name ( un nombre/descripción textual de las vías). - En la sesión de Jupyter-Lab, utilice el menú de navegación de la izquierda para navegar a la carpeta de configuraciones haciendo clic en ... y luego haciendo doble clic en configuraciones. Abra el archivo 06_Pathway_Configs.csv haciendo doble clic en él.
- Compruebe la entrada de la columna mofa_result_name y asegúrese de que corresponde al nombre del resultado MOFA generado que se utilizará para el cálculo del enriquecimiento de la vía.
- Compruebe la entrada de la columna de tipos y elimine las entradas de la columna de tipos que no contengan características que coincidan con la columna de genes del archivo Prepared_Pathway_Data.csv .
NOTA: De forma predeterminada, todas las vistas diferentes que se han utilizado en el modelo MOFA se agregan a este archivo durante la ejecución del flujo de trabajo. En caso de que haya vistas que no contengan entidades que coincidan con al menos una ruta, deben eliminarse; de lo contrario, se producirá un error en la ejecución. Un ejemplo sería que el archivo de ruta contiene solo anotaciones de ruta para genes, pero hay una vista que contiene nombres de proteínas. - Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de navegación para navegar a la carpeta de scripts haciendo clic en scripts. Abra el cuaderno 06_Downstream_Pathways.ipynb haciendo doble clic en él. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas en la parte superior, y haciendo clic en Reiniciar en la ventana emergente.
- Utilice el menú de navegación de la izquierda para navegar a la carpeta 06_figures haciendo doble clic primero en las figuras y luego en 06_figures. Abra el gráfico generado FIG06_Pathways_and_Genes_ 'mofa_result_name haciendo doble clic en él e investigue las rutas visualizadas (Figura 8C).
NOTA: La forma en que se seleccionan las rutas visualizadas se puede configurar a través del archivo de configuración. Para obtener más información, consulte la documentación de los parámetros. - Utilice el menú de navegación del lado izquierdo para navegar a la carpeta de 06_results generada haciendo clic en el símbolo de la carpeta y luego haciendo doble clic en resultados y 06_results. Compruebe si incluye el archivo que incluye las rutas enriquecidas 06_Pathway_enrichment__'mofa_result_name'.
6. Comparación de diferentes configuraciones y versiones (Figura complementaria 1, Figura complementaria 2, Figura complementaria 3, Figura complementaria 4)
- Para comparar el efecto del uso de diferentes parámetros/configuraciones a lo largo del flujo de trabajo, vuelva a ejecutar las secciones 3-5, modificando los parámetros en los archivos de configuración y utilizando diferentes identificadores de configuration_name y mofa_result_name .
NOTA: Los nuevos resultados se almacenarán con estos nombres para ser utilizados para comparar diferentes ejecuciones. - En Jupyter-Lab, use el menú de navegación de la izquierda para navegar a la carpeta de configuraciones . Abra el archivo 07_Comparison_Configs.csv haciendo doble clic en él.
- En la columna mofa_result_name , agregue los nombres de todas las ejecuciones anteriores de MOFA que se compararán (una fila por nombre/configuración, por ejemplo, MI_v1_MOFA, MI_v2_MOFA).
- En la columna compare_factors , agregue los factores que se compararán entre los modelos. De forma predeterminada, es Factor1,Factor2,Factor3. (Figura complementaria 2A).
NOTA: En este script se compararán los valores de características y factores de los diferentes modelos correlacionándolos. Esto solo funciona para los modelos que se basan en las mismas muestras (indicadas por sample_id) y el mismo conjunto de características. En caso de que las muestras o características no coincidan entre las versiones comparadas, se excluirán de la comparación. - Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de la izquierda para navegar a la carpeta de scripts haciendo clic en scripts. Abra el cuaderno 07_Compare_Models.ipynb haciendo doble clic en él. Ejecute el script haciendo clic en el botón Reiniciar kernel y ejecutar todas las celdas y haciendo clic en Reiniciar en la ventana emergente.
- Utilice el menú de la izquierda para navegar a la carpeta 06_figures haciendo doble clic primero en las figuras y luego en 06_figures. Abra los gráficos generados haciendo doble clic en los archivos para analizar la similitud de las diferentes versiones:
FIG07_Variance_Model_Comparison.pdf (Figura complementaria 2B)
FIG07_Factor_Correlations.pdf
(Figura complementaria 2C)
FIG07_Feature_Correlations.pdf
(Figura complementaria 3C)
7. Ampliación del flujo de trabajo: Adición de otros parámetros y configuraciones
NOTA: Además de los parámetros que se pueden configurar actualmente en los archivos de configuración, se pueden incluir otros ajustes en el código u otros parámetros. Por ejemplo, el propio modelo MOFA ofrece otros parámetros de entrenamiento17 que pueden modificarse directamente en el código o ajustarse a través de los archivos de configuración. En la siguiente sección del protocolo se describirá un ejemplo de cómo hacer esto para los parámetros de entrenamiento del modelo MOFA adicionales. Para esta parte, se requieren conocimientos de programación en R.
- En Jupyter-Lab, use el menú de navegación de la izquierda para navegar a la carpeta de scripts . Abra el bloc de notas 03_Run_MOFA.ipynb haciendo doble clic en él.
- Haga clic en la pestaña Tabla de contenido en el lado izquierdo y luego navegue hasta la subsección 4.3 Establecer opciones de capacitación MOFA y ejecute la capacitación modelo haciendo clic en ella. Desplácese hacia abajo para ver la salida impresa del modelo MOFA de parámetros configurables en el bloc de notas.
- Dentro del bucle R for en el código debajo del encabezado, se establecen todas las opciones de datos, modelo y entrenamiento de MOFA. Debajo de la línea model_opts$num_factors = mofa_configs$amount_of_factors[i], agregue otra línea con el código a continuación
model_opts$probabilidades['data_type'] = 'poisson'.
NOTA: Esto cambiará la distribución que el modelo toma como entrada para la vista especificada por el nombre data_type para todas las ejecuciones de MOFA. Al especificar poisson para un tipo de datos, el modelo solo se ejecutará cuando las características de este tipo de datos sean enteros (por ejemplo, recuentos de lectura de RNA-seq). Para obtener más información sobre los datos, la formación y las opciones de modelo del MOFA, también se pueden consultar los tutoriales y la documentación del MOFA17. - Guarde los cambios en el bloc de notas haciendo clic en el botón Guardar en la parte superior.
- Para entregar nuevos parámetros a través de los archivos de configuración .csv , utilice la navegación del lado izquierdo para navegar a la carpeta de configuraciones haciendo doble clic en configuraciones y abra el archivo 03_MOFA_Configs.csv haciendo doble clic.
- Agregue una nueva columna especificando el nombre del parámetro, por ejemplo, number_iterations e introduzca un valor, por ejemplo, 1000. Guarde los cambios haciendo clic en Archivo > Guardar archivo CSV.
- Utilice el menú de navegación para navegar por la carpeta de scripts haciendo clic en scripts. Abra el cuaderno 03_Run_Mofa.ipynb haciendo doble clic en él. Haga clic en la pestaña Tabla de contenido en el lado izquierdo y luego navegue hasta la subsección 4.3 Establecer opciones de capacitación MOFA y ejecute la capacitación modelo haciendo clic en ella.
- Reemplace la línea train_opts$maxiter = 50000 por train_opts$maxiter = mofa_configs$column_name[i ] (cuando el nombre de la columna agregada es number_of_iterations es train_opts$maxiter = mofa_configs$number_of_iterations[i]).
NOTA: El archivo de configuración 03_MOFA_Configs.csv en este cuaderno se lee al principio del cuaderno (subsección: Requisitos previos, configuraciones y parámetros) como mofa_config data.frame en la sesión y, por lo tanto, en esta línea de código, se hace referencia a este objeto y a la columna recién generada correspondiente. Como se pueden ejecutar varias configuraciones al mismo tiempo, i identifica la fila del data.frame a medida que la estimación del modelo se ejecuta en un bucle for en todas las diferentes filas del archivo .csv . El principio de lectura en el archivo de configuración al principio del cuaderno en la sección 'Requisitos previos, configuraciones y parámetros' es el mismo para todos los cuadernos, y se pueden realizar modificaciones adicionales como esta. - Guarde los cambios en el bloc de notas haciendo clic en el botón Guardar .
Resultados
Después de la ejecución exitosa del flujo de trabajo, se generan varias tablas y figuras como se indica en la Figura 2. Las figuras se colocan en la carpeta /figures (Figura 6, Figura 7, Figura 8, Figura complementaria 1, Figura complementaria 2, Figura complementaria 3, Figura complementaria ...
Discusión
Con el protocolo descrito, se presenta un flujo de trabajo modular y extensible basado en Jupyter-notebook que se puede utilizar para explorar rápidamente un conjunto de datos multiómico complejo. Las partes principales del flujo de trabajo consisten en la parte de preprocesamiento y armonización de datos (que ofrece diferentes pasos estándar para el filtrado y la normalización de los datos), la estimación del modelo MOFA9 y algunos ejemplos de análisis pos...
Divulgaciones
Los autores declaran no tener ningún conflicto de intereses.
Agradecimientos
C.L. cuenta con el apoyo de la Asociación Helmholtz en el marco de la escuela de investigación conjunta "Munich School for Data Science - MUDS".
Materiales
| Name | Company | Catalog Number | Comments |
| Apptainer | NA | NA | https://apptainer.org/docs/admin/main/installation.html |
| Compute server or workstation or cloud (Linux, Mac or Windows environment). Depending on the size of the different input datasets we recommend running the workflow on a suitable machine (in our setting we use: 16 CPU, 64GB Memory) | Any manufacturer | 16 CPU, 64GB Memory | Large Memory is only required for the processing of the raw single cell data. After preprocessing the later analysis steps can also be performed on regular desktop or laptop computers |
| git | NA | NA | https://git-scm.com/book/en/v2/Getting-Started-Installing-Git |
| GitHub | GitHub | NA | https://github.com/heiniglab/mofa_workflow |
Referencias
- Lähnemann, D., et al. Eleven grand challenges in single-cell data science. Genome Biol. 21 (1), 31 (2020).
- Colomé-Tatché, M., Theis, F. J. Statistical single cell multi-omics integration. Curr Opin Syst Biol. 7, 54-59 (2018).
- Hawe, J., Theis, F., Heinig, M. Inferring interaction networks from multi-omics data. Front Genet. 10, 535 (2019).
- Hawe, J. S., et al. Network reconstruction for trans acting genetic loci using multi-omics data and prior information. Genome Med. 14 (1), 125 (2022).
- Koh, H. W. L., Fermin, D., Vogel, C., Choi, K. P., Ewing, R. M., Choi, H. iOmicsPASS: network-based integration of multiomics data for predictive subnetwork discovery. NPJ Syst Biol Appl. 5, 22 (2019).
- Ogris, C., Hu, Y., Arloth, J., Müller, N. S. Versatile knowledge guided network inference method for prioritizing key regulatory factors in multi-omics data. Sci Rep. 11, 6806 (2021).
- Lee, C., vander Schaar, M. A variational information bottleneck approach to multi-omics data integration. Proceedings of The 24th International Conference on Artificial Intelligence and Statistics. 130, 1513-1521 (2021).
- Singh, A., et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 35 (17), 3055-3062 (2019).
- Argelaguet, R., et al. Multi-omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. 14 (6), e8124 (2018).
- Cantini, L., et al. Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nature Commun. 12 (1), 124 (2021).
- Pekayvaz, K., et al. Multiomic analyses uncover immunological signatures in acute and chronic coronary syndromes. Nature Medicine. 30 (6), 1696-1710 (2024).
- Swirski, F. K., Nahrendorf, M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol. 18 (12), 733-744 (2018).
- Janjic, A., et al. Prime-seq, efficient and powerful bulk RNA sequencing. Genome Biol. 23 (1), 88 (2022).
- Wolf, F. A., Angerer, P., Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19 (1), 15 (2018).
- Cao, Y., et al. Integrated analysis of multimodal single-cell data with structural similarity. Nucleic Acids Res. 50 (21), e121 (2022).
- . Get Started - JupyterLab 4.1.0a4 documentation Available from: https://jupyterlab.readthedocs.io/en/latest/getting_started/overview.html (2024)
- . MOFA2: training a model in R Available from: https://raw.githack.com/bioFAM/MOFA2_tutorials/master/R_tutorials/getting_started_R.html (2020)
- De Silva, D., et al. Robust T cell activation requires an eIF3-driven burst in T cell receptor translation. eLife. 10, e74272 (2021).
- Li, G., Liang, X., Lotze, M. HMGB1: The central cytokine for all lymphoid cells. Front Immunol. 4, 68 (2013).
- Jassal, B., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48 (D1), D498-D503 (2020).
- Argelaguet, R., et al. MOFA+: a statistical framework for comprehensive integration of multimodal single-cell data. Genome Biol. 21 (1), 111 (2020).
- Velten, B., et al. Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO. Nat Methods. 19 (2), 179-186 (2022).
- Qoku, A., Buettner, F. Encoding domain knowledge in multi-view latent variable models: A Bayesian approach with structured sparsity. Proceedings of The 26th International Conference on Artificial Intelligence and Statistics. 206, 11545-11562 (2022).
- Multi-Omics Factor Analysis. MOFA Available from: https://biofam.github.io/MOFA2/ (2024)
- Mitchel, J., et al. Tensor decomposition reveals coordinated multicellular patterns of transcriptional variation that distinguish and stratify disease individuals. bioRxiv. , (2023).
Reimpresiones y Permisos
Solicitar permiso para reutilizar el texto o las figuras de este JoVE artículos
Solicitar permisoExplorar más artículos
This article has been published
Video Coming Soon
ACERCA DE JoVE
Copyright © 2025 MyJoVE Corporation. Todos los derechos reservados