Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
A Microscopic Phenotypic Assay for the Quantification of Intracellular Mycobacteria Adapted for High-throughput/High-content Screening
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
Here, we describe a phenotypic assay applicable to the High-throughput/High-content screens of small-interfering synthetic RNA (siRNA), chemical compound, and Mycobacterium tuberculosis mutant libraries. This method relies on the detection of fluorescently labeled Mycobacterium tuberculosis within fluorescently labeled host cell using automated confocal microscopy.
Streszczenie
Despite the availability of therapy and vaccine, tuberculosis (TB) remains one of the most deadly and widespread bacterial infections in the world. Since several decades, the sudden burst of multi- and extensively-drug resistant strains is a serious threat for the control of tuberculosis. Therefore, it is essential to identify new targets and pathways critical for the causative agent of the tuberculosis, Mycobacterium tuberculosis (Mtb) and to search for novel chemicals that could become TB drugs. One approach is to set up methods suitable for the genetic and chemical screens of large scale libraries enabling the search of a needle in a haystack. To this end, we developed a phenotypic assay relying on the detection of fluorescently labeled Mtb within fluorescently labeled host cells using automated confocal microscopy. This in vitro assay allows an image based quantification of the colonization process of Mtb into the host and was optimized for the 384-well microplate format, which is proper for screens of siRNA-, chemical compound- or Mtb mutant-libraries. The images are then processed for multiparametric analysis, which provides read out inferring on the pathogenesis of Mtb within host cells.
Wprowadzenie
Among the emerging and re-emerging infectious pathogens reported during the last years, Mycobacterium tuberculosis (Mtb) holds a prominent place being responsible for 1.4 million deaths and 8.7 million new infections in 2011 (Global tuberculosis report 2012, www.who.int/topics/tuberculosis/en/). Despite the availability of multidrug therapies, the number of infected people is still on the rise and multidrug resistant (MDR) as well as extensively drug resistant (XDR) Mtb are quickly spreading all over the world1. Moreover, when taking into consideration the presence of Mtb antigens, it is evident that one third of the global population is considered as being latently infected by Mtb. Statistically, in one case out of ten, there is evolution towards the active form of the disease with subsequent clinical symptoms2. Therefore, new means to fight Mtb are urgently needed. In this context, we developed an in vitro visual phenotypic assay relying on monitoring Mtb invasion and multiplication into host cells by automated confocal fluorescence microscopy3. The adaptation of the assay in 384-well microtiter plates in combination with automated image acquisition and analysis, allowed High-content/High-throughput Screening (HC/HTS) of medium scale libraries of compounds, siRNAs and bacterial mutants. The screening of a genome wide RNAi library on this phenotypic assay thus enabled the identification of the key host-factors involved in Mtb trafficking and intracellular replication but also the elucidation of host-pathways exploited by the tubercle bacillus. Another adaptation of this particular phenotypic assay was for the identification of bacterial factors essential to Mtb intra-phagosomal persistence. For instance, the arrest of phagosome maturation is considered as one of the major mechanisms that facilitates the survival and replication of Mtb in macrophage. The monitoring of the subcellular localization of Mtb knock-out mutants in fluorescently labeled-acidic compartments allowed for the identification of bacterial genes involved in the survival process4. Finally, the high-content imaging of Mtb also offers an excellent method to quantify drug efficiency for inhibiting various phenomena like intracellular bacterial growth3. Altogether, this type of high throughput phenotypic assay allows accelerating drug discovery against TB and the data collected by these different approaches contribute to a better understanding of the host manipulation exerted by Mtb.
Protokół
1. High-throughput Genome-wide siRNA Screening
Screening performed in a human Type-II pneumocytes model A549 cell line upon infection with Mtb H37Rv expressing Green Fluorescent Protein (GFP). This procedure is outlined in Figure 1A.
- Resuspend the dried siRNA library that is stored in mother plates (96-well plates) with 1x siRNA buffer to reach a concentration of 4 μM, then transfer 10 μl of the mixture into a 384-well daughter plate (daughter plate 1).
- Add 10 μl of 1x siRNA buffer in daughter plate 1 to dilute siRNA by 2-fold. After siRNA resuspension, plates are sealed with peelable aluminum seal and can be stored at -20 °C at least 6 months and up to 2-3 years, but storage time may vary depending on siRNA library manufacturer's recommendations.
- Dilute siRNAs in daughter plate 1 into daughter plate 2 to reach a concentration of 500 nM. After siRNA resuspension, plates are sealed with peelable aluminum seal and can be stored at -20 °C at least 6 months and up to 2-3 years, but storage time may vary depending on siRNA library manufacturer's recommendations.
- Before use, thaw daughter plate 2 at room temperature.
- Take 2.5 μl of siRNA from daughter plate 2 and place into a 384-well assay plate.
- In the same 384-well assay plate in step 1.5, add 2.5 μl negative and positive control siRNA to their respective wells.
- Dilute the transfection reagent in 1x D-PBS to yield enough solution to provide 0.1 μl transfection reagent in each well and preincubate the diluted transfection solution at room temperature for 5 min.
- Add 7.5 μl of the transfection reagent/PBS solution mixture to each well in the assay plate and incubate for 30 min at room temperature.
- Add 40 μl of A549 cells (1,500 cells/well) suspended in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). Maintain cells for a 3 day-incubation period at 37 °C in an atmosphere containing 5% CO2. These cells divide every 24 hr, thus about 12,000 cells are in the wells three days after transfection.
- Wash a two-week old GFP-expressing Mtb H37Rv culture out with D-PBS (free from MgCl2 and CaCl2) by centrifugation at 4,000 x g for 5 min and discard the washes. Repeat this step 3x. (For GFP-Mtb H37Rv culture conditions see Protocol 3).
- Suspend the bacterial pellet in 10 ml of RPMI 1640 medium supplemented with 10% FBS and decant for 1 hr at room temperature to allow bacterial aggregates to sediment.
- Collect the bacterial supernatant and measure OD600 (OD600 should be between 0.6-0.8) and GFP-fluorescence (RFU value) using a microplate reader to determine the bacterial concentration. Calculate the titer of the suspension using a reference regression line displaying RFU value = f (CFU value) that had been generated prior to the experiment on another culture that had been prepared in the same conditions. Prepare bacterial suspension containing 2.4 x 106 bacteria/ml, which corresponds to a multiplicity of infection (MOI) of 5.
- Remove the medium in the 384-well assay plate and add 25 μl of freshly prepared bacterial suspension.
- Incubate the 384-well assay plate at 37 °C for 5 hr in an atmosphere containing 5% CO2.
- Remove the medium and gently wash the cells with RPMI medium supplemented with 10% FBS 3x.
- To kill the remaining extracellular bacteria, treat the cells with 50 μl of fresh RPMI-FBS medium containing 50 μg/ml of amikacin at 37 °C for 1 hr in an atmosphere containing 5% CO2.
- Remove the medium-containing amikacin and add 50 μl of fresh RPMI medium supplemented with 10% FBS. Incubate the 384-well assay plate at 37 °C for 5 days in an atmosphere containing 5% CO2.
- Prior to image acquisition, add 10 μl of freshly prepared 30 μg/ml of DAPI in PBS (final concentration 5 μg/ml) and incubate for 10 min at 37 °C.
- Load the plate into automated confocal microscope.

Click here to view larger image. - Set the exposure parameters. Record DAPI fluorescence using excitation laser 405 nm with emission filter 450 nm and GFP fluorescence using excitation laser 488 nm with emission filter 520 nm.
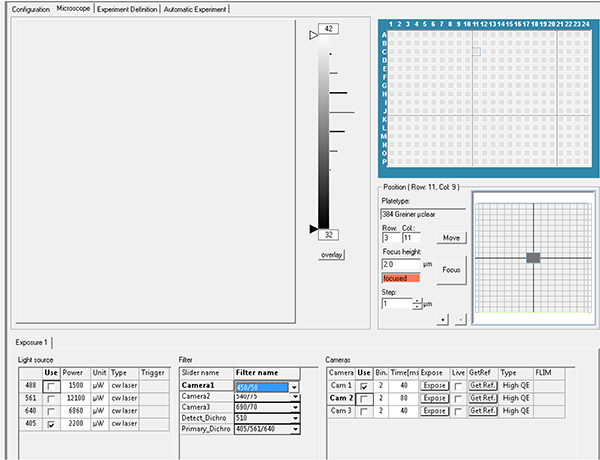
Click here to view larger image.

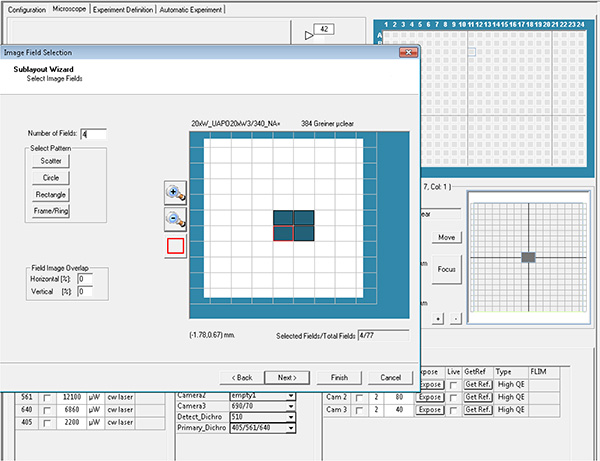
Click here to view larger image. - Select the wells and the fields in each well to be acquired, which is then referred as layout and sublayout parameters.
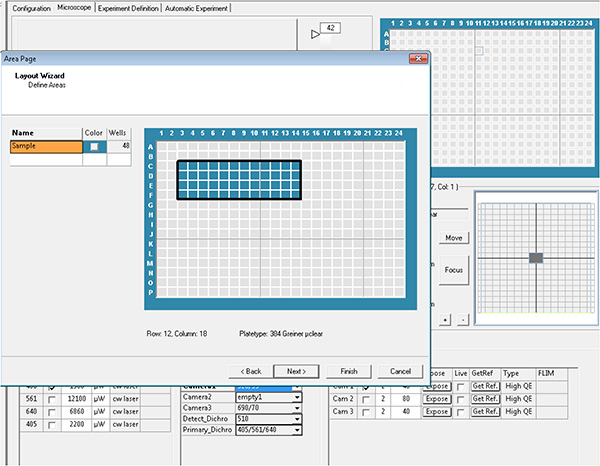
Click here to view larger image.
Click here to view larger image. - Generate the experiment file using the parameters from steps 1.20 and 1.21, and run the automatic acquisition.
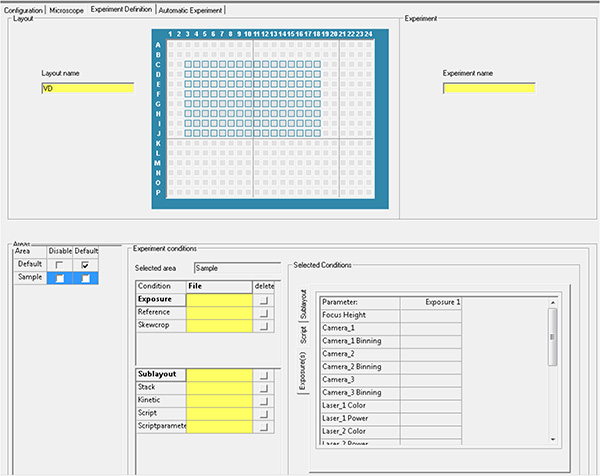
Click here to view larger image.
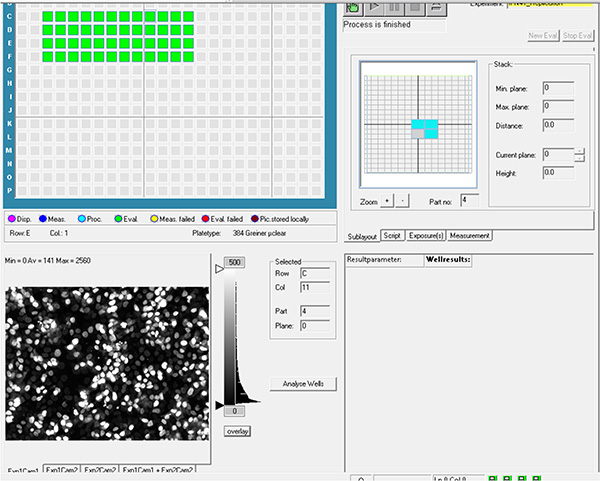
Click here to view larger image. - Transfer images to remote server.
Click here to view larger image. - Evaluate images using image analysis software. Detect cell nuclei from DAPI channel using nuclei detection algorithm and the bacterial area from GFP channel using pixel intensity properties algorithm (Figure 4A).
Note: This protocol is optimized to study the effect of gene silencing on the intracellular Mtb growth. Mtb is a slow-growth bacterium which divides every 20 hr in optimal conditions. After 5 days post-infection the amount of extracellular Mtb is still low in absence of cell lysis and didn't affect the quality of the analysis. This protocol must be optimized in terms of length of antibiotic treatment and incubation time to be adapted for siRNA screens using fast-growth bacteria like Mycobacteria smegmatis and Escherichia coli that are extensively released and can infect new cells.
2. High-throughput Compound Screening
Screening performed on Mtb H37Rv infected host cells. This procedure is outlined in Figure 1B.
- Thaw the 384-well mother plates containing the compound library solubilized in DMSO 100%. Transfer 0.5 μl of the compounds in 384-well daughter plates containing 10 μl of RPMI 1640 medium supplemented with 10% FBS.
- Wash a two-week old GFP-expressing Mtb H37Rv culture out with D-PBS (free from MgCl2 and CaCl2) by centrifugation at 4,000 x g for 5 min and discard the washes. Repeat this step 3x (For GFP-Mtb H37Rv culture conditions see Protocol 3).
- Suspend the bacterial pellet in 10 ml of RPMI 1640 medium supplemented with 10% FBS and decant for 1 hr at room temperature to allow bacterial aggregates to sediment.
- Collect the bacterial supernatant and measure OD600 (OD600 should be between 0.6-0.8) and GFP-fluorescence (RFU value) using a microplate reader. Calculate the titer of the suspension using a reference regression line displaying RFU value = f (CFU value) that had been generated prior to the experiment. Typical concentration is 1 x 108 bacteria/ml.
- Harvest 6 day old primary human macrophages at 4 x 105 cells/ml in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 50 ng/ml recombinant human M-CSF (For human Peripheral Blood Monocyte Cells purification and macrophages differentiation see Protocol 4).
- Incubate the diluted primary cells with bacilli at different MOI, ranging from 1-5, in suspension with mild shaking at 90 rpm for 2 hr at 37 °C.
- Wash the infected cells by centrifugation at 350 x g to remove the extracellular bacteria. After each centrifugation step, resuspend the pellet in RPMI 1640 medium supplemented with 10% FBS. Repeat this step 2x.
- Suspend the infected cells in RPMI 1640 medium supplemented with 10% FBS and amikacin at 50 μg/ml and incubate the suspension with mild shaking for 1 hr at 37 °C.
- Remove the cell-culture medium containing amikacin by centrifugation at 350 x g and wash the infected cells with complete RPMI 1640 medium supplemented with 10% FBS and 50 ng/ml recombinant human M-CSF. Repeat once.
- Add 40 μl of the infected macrophages suspension in the same assay plate in step 2.1, which already contains 10 μl of the compound dilutions. The final concentration of DMSO in each well has now reached 1%.
- Incubate the assay plates for 5 days at 37 °C in an atmosphere containing 5% CO2.
- Stain the live cells with cell-permeant far-red fluorescent dye.
- Load the plate into automated confocal microscope.

Click here to view larger image. - Set the exposure parameters. Record far-red fluorescence using excitation laser 640 nm with emission filter 690 nm and GFP fluorescence using excitation laser 488 nm with emission filter 520 nm.

Click here to view larger image.

Click here to view larger image. - Select the wells and the fields in each well to be acquired, which is then referred as layout and sublayout parameters.

Click here to view larger image.

Click here to view larger image. - Generate the experiment file using the parameters from steps 2.14 and 2.15 and run the automatic acquisition.

Click here to view larger image.

Click here to view larger image. - Transfer images to remote server.

Click here to view larger image. - Evaluate images using image analysis software. Detect cell area from far-red channel using pixel intensity properties algorithm and bacterial area from GFP channel using pixel intensity properties algorithm (Figure 4B).
Note: This protocol can be adapted for Mtb mutant library screening by replacing compounds by mutants expressing a fluorescent protein (One well/One mutant) (Figure 1C, see also Brodin et al.4). Fluorescent mutants are first seeded in wells (20 μl of bacterial suspension per well). Bacteria are then recovered by 30 μl of cell suspension. After centrifugation at 350 x g for 1 min, the plate is incubated at 37 °C in an atmosphere containing 5% CO2. Incubation time and MOI depend on the assay. As an example, for visualization of early cellular events such as phagosome acidification, the cells can be infected for 2 hr with MOI ranging from 1-20. Lysosomes are stained using Lysotracker dye at 2 μM for 1.5 hr at 37 °C in an atmosphere containing 5% CO2 and then fixed with either 10% formalin or 4% paraformaldehyde (PFA). Confocal images are acquired and finally analyzed using image analysis scripts featuring appropriate algorithms for lysosomes detection and subcellular localization4.
3. Green Fluorescent Protein Expressing Mycobacterium tuberculosis H37Rv (GFP-H37Rv) Culture Conditions
For long term storage, GFP-H37Rv were frozen in D-PBS (around 1 x 108 mycobacteria per vial).
- Resuspend one frozen-stock vial of GFP-H37Rv in an Erlenmeyer flask containing 50 ml of 7H9 broth medium supplemented with Middlebrook OADC enrichment 10%, Glycerol 0.5%, Tween-80 0.05%, and Hygromycin B (50 μg/ml).
- Incubate 8 days at 37 °C.
- Measure OD600 of the GFP-H37Rv culture.
- Dilute the GFP-H37Rv culture to obtain OD600 = 0.1 in fresh 7H9 broth medium supplemented with Middlebrook OADC enrichment 10%, Glycerol 0.5%, Tween-80 0.05% and Hygromycin B (50 μg/ml).
- Incubate the GFP-H37Rv at 37 °C for 8 days more before use for the assay.
4. Human Peripheral Blood Monocyte Cells Purification from Whole-blood or Buffy-coat Preparation
- Dilute the blood pouch 2x in 1x D-PBS (free from MgCl2 and CaCl2) containing 1% FBS.
- Isolate the monocytes by Ficoll density gradient centrifugation at 400 x g for 20 min.
- Collect the isolated monocytes.
- Wash the monocytes 3x with 1x D-PBS (free from MgCl2 and CaCl2) containing 1% FBS by centrifugation at 400 x g for 10 min at room temperature.
- Concentrate the cells up to 1 x 107 cell/ml.
- Purify monocytes using CD14-magnetic beads according to the manufacturer's protocol (see Materials).
- After CD14-monocytes purification, seed the cells at 1.5 x 106 cell/ml in RPMI 1640 complemented with 10% FBS and 40 ng/ml of human Macrophages Colony Stimulating Factor (hM-CSF) and incubated for 4 days at 37 °C in an atmosphere containing 5% CO2.
- After 4 days replace the medium with fresh RPMI 1640 complemented with 10% FBS and 40 ng/ml of hM-CSF and incubated for 2 days at 37 °C in an atmosphere containing 5% CO2.
- After 6 days, the cells can be used for the assay.
Wyniki
High-throughput genome-wide siRNA screening
Mtb is able to colonize immune cells in vitro as well as several other lung epithelial cells. For instance, Mtbis able to infect and damage A549 epithelial cells that are commonly used as a model for human type II pneumocytes5-7. Dectin-1 was reported as a host cell receptor involved in Mtb uptake, proinflammatory response and antibacterial effect on intracellular mycobacterial growth in A549 cells8
Dyskusje
We describe here the methods required for a phenotypic assay using a GFP-expressing Mtb H37Rv strain to infect fluorescently labeled host cells, which makes it appropriate for High-content/High-throughput screens. This protocol could be applied to a broad range of compounds, fluorescent probes and Mtb mutants. For each protocol described above, fixation and immunolabeling steps could be performed prior to image acquisition. We use an automated fluorescent confocal microscope equipped with a 20X (NA 0.70...
Ujawnienia
No conflicts of interest declared.
Podziękowania
Financial support for this work was provided by the European Community (ERC-STG INTRACELLTB Grant n° 260901, MM4TB Grant n° 260872), the Agence Nationale de Recherche, the Feder (12001407 (D-AL) Equipex Imaginex BioMed), and the Region Nord Pas de Calais. We gratefully acknowledge the technical assistance of Gaspard Deloison, Elizabeth Werkmeister, Antonino Bongiovanni and Frank Lafont from the platform BICeL.
Materiały
| Name | Company | Catalog Number | Comments |
| µclear-plate black, 384-well | Greiner Bio-One | 781091 | 127.8/86/15 MM with Lid, TC treated |
| CellCarrier 384-well plate | PerkinElmer | 6007550 | Black, Clear Bottom, with Lid, TC treated |
| V-bottom white, 384-well plate | Greiner Bio-One | 781280 | |
| sealing tape, breathable, sterile | Corning | 3345 | |
| Lipofectamine RNAiMax | Life Technologies | 13778150 | Transfection reagent |
| Dimethyl sulfoxide | Sigma-Aldrich | 34943 | |
| RPMI 1640 + GlutaMAX-I | Life Technologies | 61870-010 | Cell culture medium |
| D-PBS 1x [-]MgCl2/[-]CaCl2 | Life Technologies | 14190-094 | Dulbecco's Phosphate Salin Buffer |
| D-PBS 1x [+]MgCl2/[+]CaCl2 | Life Technologies | 14190-091 | Dulbecco's Phosphate Salin Buffer |
| Fetal bovine serum | Life Technologies | 2610040-79 | |
| Ficoll Paque PLUS | Dutscher | 17-1440-03 | Ficoll for Peripherical Blood Monocyte Cells purification |
| CD14 MicroBeads, human | Miltenyi | 130-050-201 | Purification of CD14+ Monocytes |
| Human M-CSF, premium gr. (1000 μg) | Miltenyi | 130-096-493 | Macrophage Colony Stimulating Factor |
| LS Columns | Miltenyi | 130-042-401 | Columns for CD14+ Monocytes isolation |
| Tween 80 | Euromedex | 2002-A | Mycobacteria culture |
| Glycerol high purity | Euromedex | 50405-EX | Mycobacteria culture |
| Middlebrook OADC enrichment | Becton-Dickinson | 211886 | Mycobacteria culture |
| 7H9 | Becton-Dickinson | W1701P | Mycobacteria culture |
| Versene 1x | Life Technologies | 15040033 | Nonenzymatic cell dissociation solution |
| DAPI | Life Technologies | D1306 | Nuclei dye |
| Hoechst 33342 | Life Technologies | H3570 | Nuclei dye |
| Syto60 | Life Technologies | S11342 | Nuclei/cytoplasm dye |
| Formalin | Sigma-Aldrich | HT5014 | Cell fixation solution |
| siRNA targeting Dectin-1 | Santa-Cruz | sc-63276 | |
| *siGenome* Nontargeted siRNA pool | Dharmacon | D-001206-14 | |
| Rifampicin | Sigma-Aldrich | R3501 | antibiotic |
| Isoniazid (INH) | Sigma-Aldrich | I3377-50G | antibiotic |
| Hygromycin B | Life Technologies | 10687-010 | antibiotic |
| Amikacin | Sigma-Aldrich | A1774 | antibiotic |
| Automated Confocal Microscope OPERA | PerkinElmer | Image acquisition | |
| Columbus 2.3.1 Server Database | PerkinElmer | Data transfer, storage, and analysis | |
| Acapella 2.6 software | PerkinElmer | Image-based analysis | |
| GraphPad Prism5 software | GraphPad | Statistical analysis | |
| Excel 2010 | Microsoft | Statistical analysis |
Odniesienia
- Gandhi, N. R., et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 375, 1830-1843 (2010).
- Barry, C. E., et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845-855 (2009).
- Christophe, T., Ewann, F., Jeon, H. K., Cechetto, J., Brodin, P. High-content imaging of Mycobacterium tuberculosis-infected macrophages: an in vitro model for tuberculosis drug discovery. Future Med. Chem. 2, 1283-1293 (2010).
- Brodin, P., et al. High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6, (2010).
- Bermudez, L. E., Goodman, J. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect. Immun. 64, 1400-1406 (1996).
- Dobos, K. M., Spotts, E. A., Quinn, F. D., King, C. H. Necrosis of lung epithelial cells during infection with Mycobacterium tuberculosis is preceded by cell permeation. Infect. Immun. 68, 6300-6310 (2000).
- McDonough, K. A., Kress, Y. Cytotoxicity for lung epithelial cells is a virulence-associated phenotype of Mycobacterium tuberculosis. Infect. Immun. 63, 4802-4811 (1995).
- Lee, H. M., Yuk, J. M., Shin, D. M., Jo, E. K. Dectin-1 is inducible and plays an essential role for mycobacteria-induced innate immune responses in airway epithelial cells. J. Clin. Immunol. 29, 795-805 (2009).
- Zhang, J. H., Chung, T. D., Oldenburg, K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 4, 67-73 (1999).
- Birmingham, A., et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods. 6, 569-575 (2009).
- Carralot, J. P., et al. Automated high-throughput siRNA transfection in raw 264.7 macrophages: a case study for optimization procedure. J. Biomol. Screen. 14, 151-160 (2009).
- Zhang, X. D. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J. Biomol. Screen. 16, 775-785 (2011).
- Song, O. R., et al. Confocal-based method for quantification of diffusion kinetics in microwell plates and its application for identifying a rapid mixing method for high-content/throughput screening. J. Biomol. Screen. 15, 138-147 (2010).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone