Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Kinematic Analysis of Cell Division and Expansion: Quantifying the Cellular Basis of Growth and Sampling Developmental Zones in Zea mays Leaves
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
Quantifying cell division and expansion is of crucial importance to the understanding of whole-plant growth. Here, we present a protocol to calculate cellular parameters determining maize leaf growth rates and highlight the use of these data for investigating molecular growth regulatory mechanisms by directing developmental stage-specific sampling strategies.
Streszczenie
Growth analyses are often used in plant science to investigate contrasting genotypes and the effect of environmental conditions. The cellular aspect of these analyses is of crucial importance, because growth is driven by cell division and cell elongation. Kinematic analysis represents a methodology to quantify these two processes. Moreover, this technique is easy to use in non-specialized laboratories. Here, we present a protocol for performing a kinematic analysis in monocotyledonous maize (Zea mays) leaves. Two aspects are presented: (1) the quantification of cell division and expansion parameters, and (2) the determination of the location of the developmental zones. This could serve as a basis for sampling design and/or could be useful for data interpretation of biochemical and molecular measurements with high spatial resolution in the leaf growth zone. The growth zone of maize leaves is harvested during steady-state growth. Individual leaves are used for meristem length determination using a DAPI stain and cell-length profiles using DIC microscopy. The protocol is suited for emerged monocotyledonous leaves harvested during steady-state growth, with growth zones spanning at least several centimeters. To improve the understanding of plant growth regulation, data on growth and molecular studies must be combined. Therefore, an important advantage of kinematic analysis is the possibility to correlate changes at the molecular level to well-defined stages of cellular development. Furthermore, it allows for a more focused sampling of specified developmental stages, which is useful in case of limited budget or time.
Wprowadzenie
Growth analysis depends on a set of tools that are commonly used by plant scientists to describe genotype determined growth differences and/or phenotypic responses to environmental factors. They include size and weight measurements of the whole plant or an organ and calculations of growth rates to explore the underlying mechanisms of growth. Organ growth is determined by cell division and expansion at the cellular level. Therefore, including the quantification of these two processes in growth analyses is key to understanding differences in whole-organ growth1. Consequently, it is crucial to have an appropriate methodology to determine cellular growth parameters that is relatively easy to use by non-specialized laboratories.
Kinematic analysis has already been established as an approach providing a powerful framework for the development of organ growth models2. The technique has been optimized for linear systems, such as Arabidopsis thaliana roots and monocotyledonous leaves, but also for non-linear systems, such as dicotyledonous leaves3. Nowadays, this methodology is increasingly being used to study how genetic, hormonal, developmental, and environmental factors influence cell division and expansion in various organs (Table 1). Moreover, it also provides a framework to link cellular processes to their underlying biochemical, molecular, and physiological regulations (Table 2), although limitations can be imposed by organ size and spatial organization for techniques that require higher amounts of plant material (e.g., metabolite measurements, proteomics, etc.).
Monocotyledonous leaves, such as the maize (Zea mays) leaf, represent linear systems in which cells move from the base of the leaf towards the tip, sequentially passing through the meristem and elongation zone to reach the mature zone. This makes it an ideal model system for quantitative studies of the spatial patterns of growth4. Moreover, maize leaves have large growth zones (meristem and elongation zone spanning several centimeters5) and provide possibilities for studies at other organizational levels. This allows for the investigation of the (putative) regulatory mechanisms controlling cell division and expansion, quantified by kinematic analysis through a range of molecular techniques, physiological measurements, and cell biology approaches (Table 2).
Here, we provide a protocol for performing a kinematic analysis in monocot leaves. First, we explain how to conduct a proper analysis of both cell division and cell elongation as a function of position along the leaf axis and how to calculate kinematic parameters. Secondly, we also show how this can be used as a basis for sampling design. Here, we discuss two cases: high-resolution sampling and focused sampling, enabling improved data interpretation and the saving of time/money, respectively.
Table 1. Overview of kinematic analyses methods for quantification of cell division and expansion in various organs.
| organ | reference |
| monocotyledonous leaves | 16, 20, 21, 22 |
| root tips | 2, 23, 24, 25, 26, 27, 28, 29 |
| dicotyledonous leaves | 21, 30, 31 |
| shoot apical meristem | 32 |
Table 1. Overview of kinematic analyses methods for quantification of cell division and expansion in various organs.
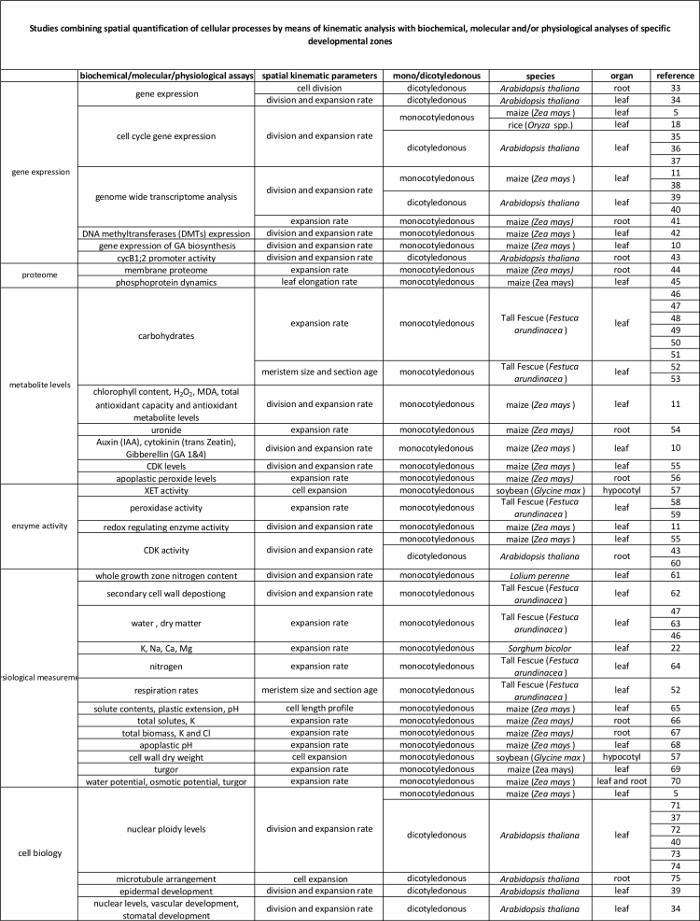
Table 2. Link between cellular processes quantified by the kinematic analysis to their regulation at the molecular level. References to various studies linking the quantification of cellular processes to results from biochemical and molecular assays in various species and organs. Xyloglucan endotransglucosylase (XET), malondialdehyde (MDA), cyclin-dependent kinases (CDK). Please click here to view a larger version of this table.
Access restricted. Please log in or start a trial to view this content.
Protokół
NOTE: The following protocol for kinematic analysis is only valid for leaves during steady-state growth. This implies a stable leaf elongation rate and spatial patterns of cell length and expansion in a leaf during a period of several days6.
1. Plant Growth and Measurements of Leaf Elongation Rate (LER)
- Choose a leaf in steady-state growth and a developmental stage of interest.
NOTE: There is a difference between steady-state growth and repetitive growth, which implies similar spatial patterns on successive leaves on the same axis. During the early stages of seedling growth, successive leaves typically grow increasingly faster due to the increasing size of the growth zone7. Although a few higher leaf positions can have a similar growth pattern8, this is a transient phase that may be affected by treatments under investigation. It is therefore important to compare lines and treatments strictly on the same leaf position, even though it may be developing at a different time. Even at a constant elongation rate, the growth rate profile is not necessarily the same at different developmental stages. Therefore, it is important to analyze leaves at the same developmental stage8, typically defined by the number of days after emergence. - To perform a full kinematic analysis of leaf growth in monocots, grow at least 15 plants for each treatment and genotype under controlled conditions in a growth room.
- At the time the leaf of interest appears (emergence from the whorl of surrounding leaves), start measuring the length of the leaf daily with a ruler until the leaf is fully expanded (Figure 1i). Leaf length implies the length from soil level to the tip of the leaf. Be careful not to break or damage the leaf, since this might alter its growth.
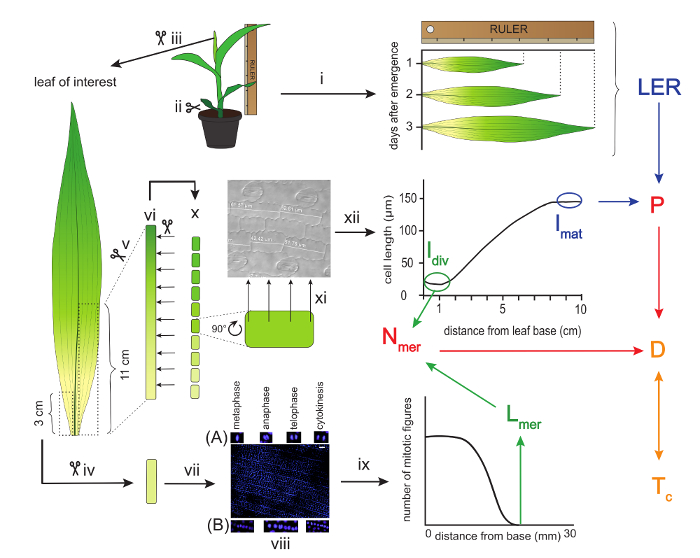
Figure 1: Schematic overview of a kinematic analysis of maize leaves. The leaf of interest is measured with a ruler for three consecutive days to calculate the Leaf Elongation Rate (LER). Thereafter, the leaf is harvested and a three-centimeter segment is used for the determination of the meristem size. This is done by measuring the length from the base up to the most distal mitotic figure after DAPI staining. (A) Examples of proliferative mitotic figures and (B) formative mitotic figures. The first eleven centimeters from the leaf base on the other side of the mid vein are used to cut ten one-centimeter segments for cell length measurements. These measurements provide the basis for creating the cell length profile, which serves to determine the mature cell length (lmat) and the length of cells leaving the meristem (ldiv). The LER and lmat are used to calculate the cell production rate (P), while ldiv and Lmer are used to calculate the number of cells in the meristem (Nmer). In turn, P and Nmer are used to calculate the average cell division rate (D), which is the inverse of cell cycle duration (Tc). Arrows of the same color indicate parameters that are used to calculate the parameter following on these arrows. Scale bars = 40 µm. Roman numbers are used to refer to specific experimental procedures described in the protocol. Please click here to view a larger version of this figure.
2. Harvesting
- At the developmental stage of interest (e.g., the third day after emergence), choose at least five representative plants from the batch on which to conduct the kinematic analysis. Continue measuring the rest of the plants as explained in step 1.3 to determine the final leaf length.
- Cut the above ground part of the plant. To keep the meristematic part intact, cut as close as possible to the roots (Figure 1ii).
- Starting from the outer leaves, remove all leaves up to the leaf of interest by gently unrolling them one by one. If necessary, remove a few extra millimeters from the base to detach the leaves. Also remove the apex and small leaves enclosed by the leaf of interest (Figure 1iii).
- Cut a 3 cm segment, starting from the base on one side of the mid vein, and store it in a 1.5 ml test tube filled with 3:1 (v:v) absolute ethanol:acetic acid solution (CAUTION: wear gloves) at 4 °C for 24 hr up to several months (Figure 1iv). This segment will later be used to determine the length of the meristem.
- From the other side of the vein, cut a 11 cm segment from the base (Figure 1 v) and place it in a 15 ml tube filled with absolute ethanol at 4 °C for at least 6 hr to remove pigments (Figure 1 vi).
NOTE: Later on, use only the first 10 cm to determine the cell length profile (see Discussion). - Renew the absolute ethanol for another round of cleaning at 4 °C for at least 24 hr (Figure 1vi).
- Finally, substitute the absolute ethanol with pure lactic acid (CAUTION: wear gloves) for cleaning and storage at 4 °C for 24 h or until further use (Figure 1vi).
3. Meristem Length Measurements
- Prepare a rinsing buffer containing 50 mM sodium chloride (NaCl), 5 mM ethylenediaminetetraacetic acid (EDTA; CAUTION: wear gloves) and 10 mM Tris(hydroxymethyl)aminomethane-hydrochloric acid (TRIS-HCl; pH 7).
- Take the 3 cm segment from section 2.4 and soak it in the buffer for 20 min (Figure 1vii).
- While waiting, use the rinsing buffer to prepare a 4',6-diamidino-2-phenylindole (DAPI) staining solution of 1 µg/ml, keeping it on ice and in the dark.
- Stain the nuclei by placing the meristem segment for 2-5 min in the DAPI staining solution. Work on ice and in the dark (Figure 1vii).
- Check for fluorescence signal by quickly mounting the segment on a microscopy glass and covering it with a cover glass. The epidermal cells should show fluorescence, while the underlying cell layers should not.
- If the staining is not sufficient, put the segment back in the DAPI staining solution for some extra minutes.
- To stop the staining, mount the segments in a drop of rinsing buffer on a microscopy slide and cover with a cover glass.
- Use a microscope equipped with UV-fluorescence at a 20X magnification, allowing for visualization of around 1,000 epidermal cells at once. Scroll throughout the segment and look for proliferative mitotic figures (metaphase, anaphase, telophase, and cytokinesis), but avoid formative cell division of the developing stomata (Figure 1viii)9. Define where the most distal mitotic figure is located.
- Determine the length of the meristem by measuring the distance between the base of the leaf and the most distal epidermal mitotic figure. Use an image-analysis software (e.g., ImageJ) to measure the full length of the image frame.
- Count the number of frames that cover the full meristem length (from the leaf base to the most distal mitotic figure) and multiply this number by the length of one frame to obtain the full meristem length (Figure 1ix).
4. Cell Length Profile
- Take the segment that is stored in lactic acid (step 2.5) and place it carefully on the bench. Cut the segments with a scalpel in 10 segments of 1 cm each (Figure 1x).
- Mount the successive leaf segments on a microscopy slide in a small drop of lactic acid. Make sure to consistently face either the adaxial or abaxial side up. In principle, there is no preference for a particular side.
- Use a microscope equipped with differential interference contrast (DIC) optics to analyze the segments, starting from the leaf base. Measure with an image analysis software the length of at least 20 replicate epidermal cells in files directly adjacent to the stomatal files in order to consistently select the same cell type.
- Do this at equally spaced positions along each of the segments (4 positions per segment suffice), and make sure to write down the corresponding position for each measurement throughout the leaf (Figure 1xi).
- Determine the average cell length at each mm along the leaf axis by using a local polynomial smoothing procedure, implemented in an R-script (Figure 1xii; Supplementary File 1).
NOTE: The R-script provides a series of data with increasing smoothing. The amount of smoothing required is somewhat arbitrary and ideally should just remove the local noise, but not affect the overall curve. Make sure to use the same amount of smoothing for all samples within one experiment. - Average the cell length at each position between plants and calculate the standard error to create a cell length profile along the leaf axis.
5. Calculations of Kinematic Parameters (See Supplementary File 2)
- Calculate the LER by taking the change in leaf length between two successive time points (e.g., 24 hr, as in step 1.3) and dividing it by the time interval.
- Calculate the length of the growth zone (Lgz) corresponding to the position distal from the base where cells reach 95% of their mature cell length on the smoothened cell length profile.
- Take for each position on the smoothened cell length profile 95% of the average of all cell lengths following that position (Figure 2).
- Compare the smoothened cell lengths (step 4.4) with the calculated 95% cell length at each position. Starting from the base of the leaf, the growth zone ends at the position where the actual cell length equals 95% of the following cell lengths (Figure 2; see Supplementary Data 2).
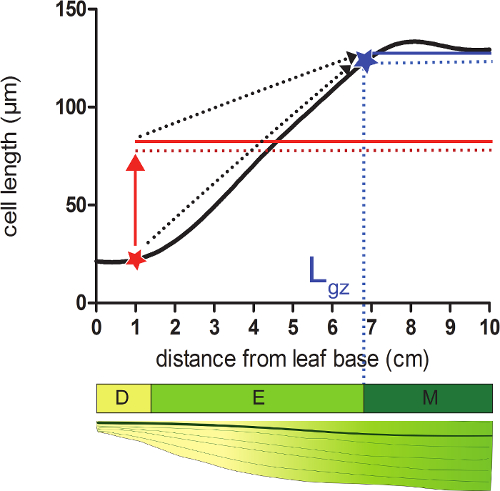
Figure 2: Determining the end of the growth zone. Meristem: At the position indicated with a red star, the actual cell size is smaller than 95% (red dotted line) of the average cell size of all cells following this position (red solid line). The end of the growth zone (Lgz; indicated with a blue star) is located where 95% (dotted blue line) of the average cell size of all cells following this position (solid blue line) equals the actual cell size. Division zone (D), elongation zone (E), and mature zone (M). Dashed arrows indicate the convergence between the local size and 95% of the average size over the distal portion of the leaf when moving from the basal positions to the tip of the leaf. Please click here to view a larger version of this figure.
- Calculate the length of elongation zone (Lel) as the difference between the length of the growth zone (Lgz) and the meristem size (Lmer; determined in step 3).
- Calculate the mature cell length (lmat) as the average cell length in the mature zone.
- Divide the LER by lmat to obtain the cell production rate (P).
- Calculate the number of cells in the elongation zone (Nel) as the difference between Ngz and Nmer. The number of cells in the meristem (Nmer) equals the cumulative number of cells located in the intervals corresponding to the meristem. The number of cells in the growth zone (Ngz) equals the cumulative number of cells located in the intervals corresponding to the growth zone.
- Calculate the average cell division rate (D) as P/Nmer. The cell cycle duration (Tc) equals ln(2)/D.
- Calculate the time in the elongation zone (Tel) by dividing Nel by P. The time in the division zone equals log2(Nmer)*Tc. The length of cells leaving the meristem (ldiv) equals the cell length from the smoothened cell length profile at the end of the meristem.
- Calculate the average cell expansion rate (Rel) using following formula: ln(lmat)-ln(ldiv)]/Tel.
Access restricted. Please log in or start a trial to view this content.
Wyniki
Here, we show a comparison between well-watered plants (control, 54% soil water content, (SWC)) and plants subjected to drought stress conditions (drought, 34% SWC) in terms of their leaf growth. All plants were grown in a growth chamber under controlled conditions (16 hr day/8 hr night, 25 °C/18 °C day/night, 300-400 µEm-2sec-1 photosynthetically active radiation (PAR). The drought conditions were established by withholding water until the correct SWC...
Access restricted. Please log in or start a trial to view this content.
Dyskusje
A full kinematic analysis on maize leaves enables the determination of the cellular basis of leaf growth and allows for the design of efficient sampling strategies. Although the protocol is relatively straightforward, some caution is recommended in the following critical steps: (1) It is important to detach the younger, enclosed leaves (step 2.3) without damaging the meristem, since meristem length determination (step 3) requires the complete meristem to be present. Some practice beforehand might be needed. (2) Meristema...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
The authors declare that they have no competing financial interests.
Podziękowania
This work was supported by a PhD fellowship from the University of Antwerp to V.A.; a PhD fellowship from the Flemish Science Foundation (FWO, 11ZI916N) to K.S.; project grants from the FWO (G0D0514N); a concerted research activity (GOA) research grant, "A Systems Biology Approach of Leaf Morphogenesis" from the research council of the University of Antwerp; and the Interuniversity Attraction Poles (IUAP VII/29, MARS), "Maize and Arabidopsis Root and Shoot Growth" from the Belgian Federal Science Policy Office (BELSPO) to G.T.S.B. Han Asard, Bulelani L. Sizani and Hamada AbdElgawad all contributed to the video.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Pots | Any | Any | We use pots with the following measures, but can be different depending on the treatment/study: bottom diameter: 11 cm, opening diameter: 15 cm, height: 12 cm. We grow one maize plant per pot. |
| Planting substrate | Any | Any | We use potting medium (Jiffy, The Netherlands), but other substrates can be used, depending on treatment/study. |
| Ruler | Any | Any | An extension ruler that covers at least 1.5 meters is needed to measure the final leaf length of the plants. |
| Seeds | Any | NA | Seeds can be ordered from a breeder. |
| Scalpel | Any | Any | The scalpel is used during leaf harvesting to detach the leaf of interest from its surrounding leaves and right after harvesting to cut a proper sample for cell length and meristem length measurements. |
| 15 mL falcon tubes | Any | Any | The 15 mL falcon tubes are used for storing samples used for cell length measurements during sample clearing with absolute ethanol and lactic acid. |
| Eppendorf tubes | Any | Any | The eppendorf tubes are used for storing samples used for meristem length measurements in ethanol:acetic acid 3:1 (v:v) solution. |
| Gloves | Any | Any | Latex gloves, which protect against corrosive reagents. |
| Acetic acid | Any | Any | CAUTION: Corrosive to metals, category 1 Skin corrosion, categories 1A,1B,1C Serious eye damage, category 1; Flammable liquids, categories 1,2,3 |
| Absolute ethanol | Any | Any | CAUTION: Hazardous in case of skin contact (irritant), of eye contact (irritant), of inhalation. Slightly hazardous in case of skin contact (permeator), of ingestion |
| Lactic acid >98% | Any | Any | CAUTION: Corrosive to metals, category 1 Skin corrosion, categories 1A,1B,1C Serious eye damage, category 1 |
| Sodium chloride (NaCl) | Any | Any | |
| Ethylenediaminetetraacetic acid (EDTA) | Any | Any | CAUTION: Acute toxicity (oral, dermal, inhalation), category 4 Skin irritation, category 2 Eye irritation, category 2 Skin sensitisation, category 1 Specific Target Organ Toxicity – Single exposure, category 3 |
| Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) | Any | Any | This material can be an irritant, contact with eyes and skin should be avoided. Inhalation of dust may be irritating to the respiratory tract. |
| 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI) | Any | Any | Cell permeable fluorescent minor groove-binding probe for DNA. Causes skin irritation. May cause an allergic skin reaction. May cause respiratory irritation. |
| Ice | Any | NA | The DAPI solution has to be kept on ice. |
| Fluorescent microscope | AxioScope A1, Axiocam ICm1 from Zeiss or other | Any fluorescent microscope can be used for determining meristem length. | |
| Microscopic slide | Any | Any | |
| Cover glass | Any | Any | |
| Tweezers | Any | Any | Tweezers are needed for unfolding the rolled maize leaf right after harvesting in order to cut a proper sample for cell length and meristem length measurements. |
| Image-analysis software | Axiovision (Release 4.8) from Zeiss | NA | The software can be downloaded at: http://www.zeiss.com/microscopy/en_de/downloads/axiovision.html. Other softwares such as ImageJ (https://imagej.nih.gov/ij/) could be used as well. |
| Microscope equipped with DIC | AxioScope A1, Axiocam ICm1 from Zeiss or other | Any microscope, equipped with differential interference contrast (DIC) can be used to measure cell lengths. | |
| R statistical analysis software | R Foundation for Statistical Computing | NA | Open source; Could be downloaded at https://www.r-project.org/ |
| R script | NA | NA | We use the kernel smoothing function locpoly of the Kern Smooth package (Wand MP, Jones MC. Kernel Smoothing: Chapman & Hall/CRC (1995)). The script is available for Mac and Windows upon inquiry with the corresponding author. |
Odniesienia
- Fiorani, F., Beemster, G. T. S. Quantitative analyses of cell division in plants. Plant Mol. Biol. 60, 963-979 (2006).
- Silk, W. K., Erickson, R. O. Kinematics of Plant-Growth. J. Theor. Biol. 76, 481-501 (1979).
- Rymen, B., Coppens, F., Dhondt, S., Fiorani, F., Beemster, G. T. S. Kinematic Analysis of Cell Division and Expansion. Plant Developmental Biology. Hennig, L., Köhler, C. , Chapter 14 (2010).
- Avramova, V., Sprangers, K., Beemster, G. T. S. The Maize Leaf: Another Perspective on Growth Regulation. Trends Plant Sci. 20, 787-797 (2015).
- Rymen, B., et al. Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol. 143, 1429-1438 (2007).
- Muller, B., Reymond, M., Tardieu, F. The elongation rate at the base of a maize leaf shows an invariant pattern during both the steady-state elongation and the establishment of the elongation zone. J. Exp. Bot. 52, 1259-1268 (2001).
- Beemster, G. T. S., Masle, J., Williamson, R. E., Farquhar, G. D. Effects of soil resistance to root penetration on leaf expansion in wheat (Triticum aestivum L): Kinematic analysis of leaf elongation. J. Exp. Bot. 47, 1663-1678 (1996).
- Bernstein, N., Silk, W. K., Lauchli, A. Growth and Development of Sorghum Leaves under Conditions of Nacl Stress - Spatial and Temporal Aspects of Leaf Growth-Inhibition. Planta. 191, 433-439 (1993).
- Sylvester, A. W., Smith, L. G. Cell Biology of Maize Leaf Development. Handbook of maize: It's Biology. Bennetzen, J. L., Hake, S. C. , Springer. NY. (2009).
- Nelissen, H., et al. A Local Maximum in Gibberellin Levels Regulates Maize Leaf Growth by Spatial Control of Cell Division. Curr. Biol. 22, 1183-1187 (2012).
- Avramova, V., et al. Drought Induces Distinct Growth Response, Protection, and Recovery Mechanisms in the Maize Leaf Growth Zone. Plant Physiol. 169, 1382-1396 (2015).
- Picaud, J. C., et al. Total malondialdehyde (MDA) concentrations as a marker of lipid peroxidation in all-in-one parenteral nutrition admixtures (APA) used in newborn infants. Pediatr. Res. 53, 406(2003).
- Basu, P., Pal, A., Lynch, J. P., Brown, K. M. A novel image-analysis technique for kinematic study of growth and curvature. Plant Physiol. 145, 305-316 (2007).
- Vander Weele, C. M., et al. A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth. Roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol. 132, 1138-1148 (2003).
- Nelissen, H., Rymen, B., Coppens, F., Dhondt, S., Fiorani, F., Beemster, G. T. S. Plant Organogenesis. DeSmet, I. , Chapter 17 (2013).
- Ben-Haj-Salah, H., Tardieu, F. Temperature Affects Expansion Rate of Maize Leaves without Change in Spatial-Distribution of Cell Length - Analysis of the Coordination between Cell-Division and Cell Expansion. Plant Physiol. 109, 861-870 (1995).
- Fiorani, F., Beemster, G. T. S., Bultynck, L., Lambers, H. Can meristematic activity determine variation in leaf size and elongation rate among four Poa species? A kinematic study. Plant Physiol. 124, 845-855 (2000).
- Pettko-Szandtner, A., et al. Core cell cycle regulatory genes in rice and their expression profiles across the growth zone of the leaf. J. Plant Res. 128, 953-974 (2015).
- Poorter, H., Remkes, C. Leaf-Area Ratio and Net Assimilation Rate of 24 Wild-Species Differing in Relative Growth-Rate. Oecologia. 83, 553-559 (1990).
- Macadam, J. W., Volenec, J. J., Nelson, C. J. Effects of Nitrogen on Mesophyll Cell-Division and Epidermal-Cell Elongation in Tall Fescue Leaf Blades. Plant Physiol. 89, 549-556 (1989).
- Tardieu, F., Granier, C. Quantitative analysis of cell division in leaves: methods, developmental patterns and effects of environmental conditions. Plant Mol. Biol. 43, 555-567 (2000).
- Bernstein, N., Silk, W. K., Lauchli, A. Growth and Development of Sorghum Leaves under Conditions of Nacl Stress - Possible Role of Some Mineral Elements in Growth-Inhibition. Planta. 196, 699-705 (1995).
- Erickson, R. O., Sax, K. B. Rates of Cell-Division and Cell Elongation in the Growth of the Primary Root of Zea-Mays. P. Am. Philos. Soc. 100, 499-514 (1956).
- Beemster, G. T. S., Baskin, T. I. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515-1526 (1998).
- Goodwin, R. H., Stepka, W. Growth and differentiation in the root tip of Phleum pratense. Am. J. Bot. 32, 36-46 (1945).
- Hejnowicz, Z. Growth and Cell Division in the Apical Meristem of Wheat Roots. Physiologia Plantarum. 12, 124-138 (1959).
- Gandar, P. W. Growth in Root Apices .1. The Kinematic Description of Growth. Bot. Gaz. 144, 1-10 (1983).
- Baskin, T. I., Cork, A., Williamson, R. E., Gorst, J. R. Stunted-Plant-1, a Gene Required for Expansion in Rapidly Elongating but Not in Dividing Cells and Mediating Root-Growth Responses to Applied Cytokinin. Plant Physiol. 107, 233-243 (1995).
- Sacks, M. M., Silk, W. K., Burman, P. Effect of water stress on cortical cell division rates within the apical meristem of primary roots of maize. Plant Physiol. 114, 519-527 (1997).
- Granier, C., Tardieu, F. Spatial and temporal analyses of expansion and cell cycle in sunflower leaves - A common pattern of development for all zones of a leaf and different leaves of a plant. Plant Physiol. 116, 991-1001 (1998).
- De Veylder, L., et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 13, 1653-1667 (2001).
- Kwiatkowska, D. Surface growth at the reproductive shoot apex of Arabidopsis thaliana pin-formed 1 and wild type. J. Exp. Bot. 55, 1021-1032 (2004).
- Kutschmar, A., et al. PSK-alpha promotes root growth in Arabidopsis. New Phytol. 181, 820-831 (2009).
- Vanneste, S., et al. Plant CYCA2s are G2/M regulators that are transcriptionally repressed during differentiation. Embo J. 30, 3430-3441 (2011).
- Eloy, N. B., et al. Functional Analysis of the anaphase-Promoting Complex Subunit 10. Plant J. 68, 553-563 (2011).
- Eloy, N. B., et al. SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. P. Natl. Acad. Sci. USA. 109, 13853-13858 (2012).
- Dhondt, S., et al. SHORT-ROOT and SCARECROW Regulate Leaf Growth in Arabidopsis by Stimulating S-Phase Progression of the Cell Cycle. Plant Physiol. 154, 1183-1195 (2010).
- Baute, J., et al. Correlation analysis of the transcriptome of growing leaves with mature leaf parameters in a maize RIL population. Genome Biol. 16, (2015).
- Andriankaja, M., et al. Exit from Proliferation during Leaf Development in Arabidopsis thaliana: A Not-So-Gradual Process. Dev. Cell. 22, 64-78 (2012).
- Beemster, G. T. S., et al. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138, 734-743 (2005).
- Spollen, W. G., et al. Spatial distribution of transcript changes in the maize primary root elongation zone at low water potential. Bmc Plant Biol. 8, (2008).
- Candaele, J., et al. Differential Methylation during Maize Leaf Growth Targets Developmentally Regulated Genes. Plant Physiol. 164, 1350-1364 (2014).
- West, G., Inze, D., Beemster, G. T. S. Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135, 1050-1058 (2004).
- Zhang, Z., Voothuluru, P., Yamaguchi, M., Sharp, R. E., Peck, S. C. Developmental distribution of the plasma membrane-enriched proteome in the maize primary root growth zone. Front. Plant Sci. 4, (2013).
- Bonhomme, L., Valot, B., Tardieu, F., Zivy, M. Phosphoproteome Dynamics Upon Changes in Plant Water Status Reveal Early Events Associated With Rapid Growth Adjustment in Maize Leaves. Mol. Cell Proteomics. 11, 957-972 (2012).
- Schnyder, H., Nelson, C. J. Growth-Rates and Assimilate Partitioning in the Elongation Zone of Tall Fescue Leaf Blades at High and Low Irradiance. Plant Physiol. 90, 1201-1206 (1989).
- Schnyder, H., Nelson, C. J., Spollen, W. G. Diurnal Growth of Tall Fescue Leaf Blades .2. Dry-Matter Partitioning and Carbohydrate-Metabolism in the Elongation Zone and Adjacent Expanded Tissue. Plant Physiol. 86, 1077-1083 (1988).
- Schnyder, H., Nelson, C. J. Growth-Rates and Carbohydrate Fluxes within the Elongation Zone of Tall Fescue Leaf Blades. Plant Physiol. 85, 548-553 (1987).
- Vassey, T. L., Shnyder, H. S., Spollen, W. G., Nelson, C. J. Cellular Characterisation and Fructan Profiles in Expanding Tall Fescue. Curr. T. Pl. B. 4, 227-229 (1985).
- Allard, G., Nelson, C. J. Photosynthate Partitioning in Basal Zones of Tall Fescue Leaf Blades. Plant Physiol. 95, 663-668 (1991).
- Spollen, W. G., Nelson, C. J. Response of Fructan to Water-Deficit in Growing Leaves of Tall Fescue. Plant Physiol. 106, 329-336 (1994).
- Volenec, J. J., Nelson, C. J. Carbohydrate-Metabolism in Leaf Meristems of Tall Fescue .1. Relationship to Genetically Altered Leaf Elongation Rates. Plant Physiol. 74, 590-594 (1984).
- Volenec, J. J., Nelson, C. J. Carbohydrate-Metabolism in Leaf Meristems of Tall Fescue .2. Relationship to Leaf Elongation Rates Modified by Nitrogen-Fertilization. Plant Physiol. 74, 595-600 (1984).
- Silk, W. K., Walker, R. C., Labavitch, J. Uronide Deposition Rates in the Primary Root of Zea-Mays. Plant Physiol. 74, 721-726 (1984).
- Granier, C., Inze, D., Tardieu, F. Spatial distribution of cell division rate can be deduced from that of p34(cdc2) kinase activity in maize leaves grown at contrasting temperatures and soil water conditions. Plant Physiol. 124, 1393-1402 (2000).
- Voothuluru, P., Sharp, R. E. Apoplastic hydrogen peroxide in the growth zone of the maize primary root under water stress.1. Increased levels are specific to the apical region of growth maintenance. J. Exp. Bot. 64, 1223-1233 (2012).
- Wu, Y. J., Jeong, B. R., Fry, S. C., Boyer, J. S. Change in XET activities, cell wall extensibility and hypocotyl elongation of soybean seedlings at low water potential. Planta. 220, 593-601 (2005).
- Macadam, J. W., Nelson, C. J., Sharp, R. E. Peroxidase-Activity in the Leaf Elongation Zone of Tall Fescue .1. Spatial-Distribution of Ionically Bound Peroxidase-Activity in Genotypes Differing in Length of the Elongation Zone. Plant Physiol. 99, 872-878 (1992).
- Macadam, J. W., Sharp, R. E., Nelson, C. J. Peroxidase-Activity in the Leaf Elongation Zone of Tall Fescue .2. Spatial-Distribution of Apoplastic Peroxidase-Activity in Genotypes Differing in Length of the Elongation Zone. Plant Physiol. 99, 879-885 (1992).
- Beemster, G. T. S., De Vusser, K., De Tavernier, E., De Bock, K., Inze, D. Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 129, 854-864 (2002).
- Kavanova, M., Lattanzi, F. A., Schnyder, H. Nitrogen deficiency inhibits leaf blade growth in Lolium perenne by increasing cell cycle duration and decreasing mitotic and post-mitotic growth rates. Plant Cell Environ. 31, 727-737 (2008).
- Macadam, J. W., Nelson, C. J. Secondary cell wall deposition causes radial growth of fibre cells in the maturation zone of elongating tall fescue leaf blades. Ann. Bot-London. 89, 89-96 (2002).
- Schnyder, H., Nelson, C. J. Diurnal Growth of Tall Fescue Leaf Blades .1. Spatial-Distribution of Growth, Deposition of Water, and Assimilate Import in the Elongation Zone. Plant Physiol. 86, 1070-1076 (1988).
- Gastal, F., Nelson, C. J. Nitrogen Use within the Growing Leaf Blade of Tall Fescue. Plant Physiol. 105, 191-197 (1994).
- Vanvolkenburgh, E., Boyer, J. S. Inhibitory Effects of Water Deficit on Maize Leaf Elongation. Plant Physiol. 77, 190-194 (1985).
- Silk, W. K., Hsiao, T. C., Diedenhofen, U., Matson, C. Spatial Distributions of Potassium, Solutes, and Their Deposition Rates in the Growth Zone of the Primary Corn Root. Plant Physiol. 82, 853-858 (1986).
- Meiri, A., Silk, W. K., Lauchli, A. Growth and Deposition of Inorganic Nutrient Elements in Developing Leaves of Zea-Mays L. Plant Physiol. 99, 972-978 (1992).
- Neves-Piestun, B. G., Bernstein, N. Salinity-induced inhibition of leaf elongation in maize is not mediated by changes in cell wall acidification capacity. Plant Physiol. 125, 1419-1428 (2001).
- Bouchabke, O., Tardieu, F., Simonneau, T. Leaf growth and turgor in growing cells of maize (Zea mays L.) respond to evaporative demand under moderate irrigation but not in water-saturated soil. Plant Cell Environ. 29, 1138-1148 (2006).
- Westgate, M. E., Boyer, J. S. Transpiration-Induced and Growth-Induced Water Potentials in Maize. Plant Physiol. 74, 882-889 (1984).
- Horiguchi, G., Gonzalez, N., Beemster, G. T. S., Inze, D., Tsukaya, H. Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana. Plant J. 60, 122-133 (2009).
- Fleury, D., et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell. 19, 417-432 (2007).
- Vlieghe, K., et al. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15, 59-63 (2005).
- Boudolf, V., et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 16, 2683-2692 (2004).
- Baskin, T. I., Beemster, G. T. S., Judy-March, J. E., Marga, F. Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135, 2279-2290 (2004).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone