A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Pulse-chase Analysis of N-linked Sugar Chains from Glycoproteins in Mammalian Cells
In This Article
Summary
We describe a method for analysis of the alteration of N-linked glycans through the early life of glycoproteins after their biosynthesis in mammalian cells. This is achieved by pulse-chase analysis of metabolically labeled glycans, enzymatic release from glycoproteins and examination by HPLC.
Abstract
Protocol
The following protocol is intended for analysis of sugar chains of total glycoproteins or of a specific glycoprotein of interest. The procedure is basically the same for both cases with a few alterations as stated throughout the protocol.
- For metabolic labeling of N-linked glycans one needs to use a subconfluent culture of mammalian cells grown overnight in a 90 mm dish for each sample (samples may represent different time points or treatments). This protocol is optimized for NIH 3T3 cells.
- Starve the cells for glucose by incubation in 5 ml of freshly prepared pre-warmed (37°C) glucose-free medium, supplied with 10% dialyzed FCS and 4 mM sodium pyruvate for 5-15 minutes.
- Replace the starvation medium with 1 ml of pre-warmed (37°C) glucose-free medium containing 400 μCi of [2-3H]-labeled mannose (lower concentration of 65 μCi/ml is used for labeling of total glycoproteins), and incubate the cells in the normal tissue culture conditions for 1 hour.
- Remove the labeling medium from each sample, and carefully add 2 ml of PBS at 4°C to the samples corresponding to pulse and place them on ice (these samples are referred to as pulses), while the samples corresponding to the chase need to be rinsed 3 times with 2 ml of pre-warmed (37°C) normal complete culture medium, and then placed in a CO2 incubator at 37°C with 5 ml of pre-warmed regular medium for the desired chase periods.
- Rinse the pulse samples (previously placed on ice) 3 times with 2 ml of ice-cold PBS. The cells are then scraped off the dish in 2 ml PBS using a cell scraper, and placed into a 2 ml Eppendorf tube.
- Short-spin the cells at 16000Xg (6-10 sec), discard the supernatant and freeze the cell pellet at -80°C.
- Perform steps 5 and 6 also for the chase samples at the end of the chase time.
- Remove the cells from the freezer and lyse them by addition of 300 μl of buffer A, vortexing briefly and incubating for 20 min on ice, or in the case of total glycoproteins analysis, by incubation 20 min on ice with buffer B, followed by sonication (4 times, 10 sec, maximum amplitude), then boil the samples for 5min.
- Centrifuge the lysates at 16000Xg for 20 min at 4°C. Transfer the supernatant to a new Eppendorf tube and discard the pellet.
- If analyzing total glycoproteins, proceed to molecular filtration and deglycosylation (step 17). For immunoprecipitation of the H2a glycoprotein demonstrated here, add 20l/sample of (Repligen) protein A-agarose beads 1:1 suspension and 3 μl/sample of our rabbit polyclonal anti H2a antibody for the desired glycoprotein (in our example anti-H2a antibody), to the supernatant from step 9.
- Incubate the samples for 4-16 hours at 4°C, constantly mixing by slow rotation.
- Spin down the beads at 16000Xg for 30 seconds, and carefully remove the supernatant using vacuum.
- Rinse the beads, by adding 500 μl buffer D and vortexing. Spin as in step 12 and remove the supernatant. Repeat the wash 3 times.
- Add 10μl denaturing buffer (supplied with the NEB endo-H kit) to the bead pellet and boil the samples for 5 min (when total proteins are analyzed, the deglycosylation is performed on the Microcon filter, step 17).
- Spin down the beads (16000Xg for 30 seconds), and transfer the supernatant into a new Eppendorf tube. Then 0.5μl of endo H enzyme are added to each sample along with 10 μl of the reaction buffer (also supplied with the NEB endo H kit), and the samples are incubated at 37°C for 3 hours.
- To separate the released glycans from the proteins, the sample is diluted 5 times with deionized water and placed on top of a molecular filter (Microcon Ultracel YM30), with a 30kDa cut-off, then centrifuged at 14000Xg for 3 min. Apply 50μl of deionized water and repeat centrifugation of the samples. This washout is repeated 2 more times, keeping the flow-through.
- If you are analyzing total glycoproteins, apply the supernatant of the lysates (from step 9) to the Microcon filter. Wash the extract with 100 μl of buffer E, 3 times by repeated centrifugation at 14000Xg for 3 min. Add 3 μl of 10X endo H reaction buffer and 1.5 μl of the endo H enzyme to the Microcon retentate, and incubate for 3 hours at 37°C. The released glycans are eluted with deionized water by 3 repeated centrifugations, as in step 16.
- If desired, retentates from steps 16 and 17, containing endo H-resistant glycoproteins are then incubated with 200mU of N-glycosidase F in 15 μl of buffer C, at 37°C for 16 h. Elution with deionized water is done as in step 16.
- Place the tube containing the endo H reaction flow-through (steps 16 and 17) in a SpeedVac concentrator, and dry the samples completely (this can be done up to 45°C to accelerate this process).
- To prepare the samples for the HPLC, resuspend the dry pellets in 12 μl of the HPLC solvent (acetonitrile:water, 60:40, 1% phosphoric acid).
- Adjust the HPLC device (connected to the Spherisorb column) to constant flow of the solvent (1 ml/min) and pressure (a particular value between 1000 and 2000 psi), and place a fraction collector to change a tube every 1 minute.
- Load the samples into the HPLC device and start the fraction collector simultaneously.
- Collect 48 fractions of 1 ml from each sample run and from a run of a standard oligosaccharide mixture.
- Transfer 500 μl from each fraction to a scintillation vial, and mix the contents with 3 ml of water-miscible scintillation fluid.
- Load the vials and read in a scintillation counter.
- The cpm readout is plotted as a function of fraction number.
- The cpm values within a peak represent a specific glycan species as follows: fractions 18-21 correspond to M5, fractions 22-26 to M6, fractions 27-31 to M7, fractions 32-35 to M8, fractions 36-39 to M9, fractions 40-42 to G1M9 and fractions 43-45 to G2M9.
- The sum of cpm values for the fractions within each peak reflects the absolute amount of a specific glycan species. In order to compensate for the fact that species containing more mannose residues acquire more label, the "relative molar amount" of each glycan species is calculated as follows: the absolute amount of each glycan species is divided by the number of mannose residues that it contains. This value is then divided by the sum of the relative molar amounts of all glycan species obtained to obtain the "percent of total" for each specific glycan species.
| Chase (h) | 0 | 4 |
| Released with endo H (cpm) a | 90000 | 16800 |
| Released with N-glycosidase F (cpm) b | 20400 | 3900 |
| Glycoproteins in retentate (cpm) c | 371700 | 127695 |
| Dolichol-oligosaccharides in retentate (cpm) d | 4350 | 540 |
| Dolichol-oligosaccharides in retentate (%) e | 1.4 ± 0.7 | 0.2 ± 0.2 |
Table 1. Analysis of release of N-linked sugar chains with deglycosidases and of the presence of dolichol-oligosaccharides in glycoprotein samples.
We applied the pulse-chase procedure to a lysate from NIH 3T3 cells. After Microcon filtration, labeled N-linked oligosaccharides were released mainly with endo H after pulse labeling (high mannose type) and with further treatment with N-glycosidase F after a chase period, which would correspond to complex type (Golgi-processed glycans), endo H resistant oligosaccharides. This quantitative analysis determined that dolichol-oligosaccharides, accounted for an insignificant fraction of the label in the initial retentate.
Notes:
- NIH 3T3 cells were pulse labeled with [2-3H] mannose and chased with complete medium. After cell lysis and filtration through Microcon YM30, high mannose N-linked oligosaccharides were released from retentates by incubation with endo H.
- Endo H-resistant material in Microcon retentates from (a) was then incubated with N-glycosidase F and released oligosaccharides measured in a scintillation counter.
- Microcon retentates obtained as in (a) before endo H treatment, were extracted 4 times with chloroform:methanol:water 10:10:3 to remove precursor glycolipids, and the unextracted material solubilized in 1N NaOH and counted in a scintillation counter.
- Dolichol-oligosaccharides were purified from extracts from (c) as described in 1 and counted in a scintillation counter.
- Average of 3 experiments for percent of dolichol-oligosaccharides (d) of total cpm in retentate (c+d).
Representative Results
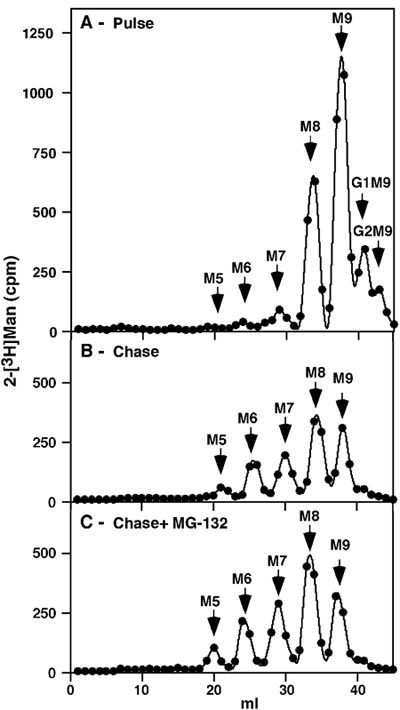
Figure 1. Pulse-chase analysis of total NIH 3T3 glycoproteins in untreated cells and upon proteasomal inhibition. After 1h pulse-labeling with [2-3H] we obtained an expected profile with some glucosylated precursors remaining (Glc2Man9GlcNAc2 (G2M9) and G1M9), but most of the label present in M9 and M8 species, the latter being the result of trimming of all the glucose and one mannose residue (Figure 1 A). No free mannose or other small precursors were detected, attesting to the thoroughness of the purification. Following a rather long 8h chase there was more extensive trimming to M7-6 and a small amount of M5, but the major species continued to be M9 and M8 (Figure 1 B). If the chase was done in the presence of a proteasomal inhibitor (30 μM MG-132), there was only a minor accumulation of these same trimmed species (Figure 1 C).
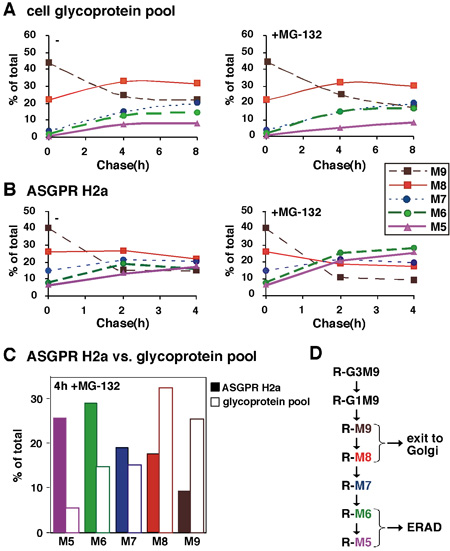
Figure 2. Quantitative analysis of sugar chains of an ERAD substrate compared to total glycoproteins. (A) In an experiment similar to the one in Figure 1, relative molar amounts of each oligosaccharide species were calculated based on mannose content. We converted the cpm values obtained for each glycan species, the percent of each species relative to the total sum of the relative molar amounts of all species present was then plotted as a function of chase time for an average of two experiments. (B) Similar to (A) except that glycans released from the ERAD substrate ASGPR H2a were analyzed after the pulse labeling and chase for up to 4h, in the presence or absence of the proteasomal inhibitor MG-132. (C) Values for 4h chase in the presence of MG-132 from (A) and (B) are compared for ASGPR H2a and the glycoprotein pool. (D) Scheme of N-linked sugar chain trimming processes in the ER. Sugar trimming processes that lead to M6-5 linked to proteins (R) that are targeted to ERAD compared to M9-8 on those that exit to the Golgi and beyond. The results indicate that the ERAD process is associated with the mannose trimming of N-glycans to yield species with 5-6 mannose residues remaining.
Discussion
The pulse-chase analysis of glycans in live cells with HPLC separation provides a method for studying the dynamics of oligosaccharide structural alterations throughout the life of a glycoprotein. There is growing evidence that such alterations are involved in producing the signals for ER folding, quality control and trafficking systems 2-5. The method can be applied not only for a specific glycoprotein of interest but also for analyzing the dynamics of the structure of total glycoproteins, as illustrated in th...
Acknowledgements
We thank Zehavit Frenkel and Sandra Tolchinsky for technical assistance. Research related to this work is supported by grants from the Israel Science Foundation (1229/07) and German-Israeli Project Cooperation (DIP-DFG).
Materials
| Name | Company | Catalog Number | Comments |
| 600E Multisolvent delivery System controller | Waters | WAT062710 | |
| Acetonitrile LiChrosolv (gradient grade for liquid chromatography) | Merck & Co., Inc. | 1.0003 | |
| Microcon Amicon Ultra 0.5 ml 30K or Centricon ultracel YM-30 | EMD Millipore | UFC503024 or 4208 | |
| Concentrator 5301, incl. 48 x 1.5 / 2.0 ml fixed-angle rotor | Eppendorf | 5301 000.016 | |
| Dialyzed F–tal Calf Serum | Biological Industries | 04-011-1A | |
| Dulbecco’s Modified Eagle’s Medium | GIBCO, by Life Technologies | 41965039 | |
| Dulbecco’s Modified Eagle’s Medium - glucose free | Sigma-Aldrich | D5030 | |
| Dulbecco’s Phosphate Buffered Saline (PBS) | Sigma-Aldrich | D1408 | |
| EDTA disodium salt(Titriplex Iii) | Merck & Co., Inc. | 1084180250 | |
| Endo Hf Kit (1,000,000 units/ml) | New England Biolabs | p0703S | |
| F–tal Calf Serum (FCS) | Biological Industries | 04-001-1A | |
| Frac-100 fraction collector | Amersham | 18-1000-77 | |
| LS 6500 Liquid Scintillation Counting systems | Beckman Coulter Inc. | 510720 | |
| Mannose, D-[2-3H(N)]- (Specific Activity: 15-30Ci/mmol) | PerkinElmer, Inc. | NET570A | |
| N-carbobenzoxyl-leucinyl-leucinyl-leucinal (MG-132) | Calbiochem | 474790 | |
| N-glycosidase F (1000 units/ml) | Roche Group | 11365177 | |
| N-octylglucoside | Sigma-Aldrich | O3757 | |
| NIH 3T3 cells | American Type Culture Collection | CRL-1658 | |
| Opti-Flour | PerkinElmer, Inc. | 6013199 | |
| Phosphoric acid solution ( 49-51%, for HPLC) | Fluka | 79607 | |
| Protease Inhibitor Cocktail | Sigma-Aldrich | P2714 | |
| Protein A-Sepharose beads | Repligen | IPA300 | |
| Rabbit polyclonal anti-H2 carboxy-terminal 6 | |||
| Sodium deoxycholate | Sigma-Aldrich | 30970 | |
| Sodium dodecyl sulfate | Bio-Rad | 161-0301 | |
| Sodium phosphate | Sigma-Aldrich | 342483 | |
| Sodium pyruvate solution (100mM) | Sigma-Aldrich | S8636 | |
| Spherisorb NH2 Column, 5 μm, 4.6 x 250 mm | Waters | PSS831115 | |
| Triton X-100 | VWR | 306324N | |
| Vibra-Cell ultrasonic processors VCX 750 | Sonics and Materials, Inc. | 690-003 | |
| Buffer A: 1% Triton X-100, 0.5% w/v sodium deoxycholate, Protease inhibitor cocktail 2% v/v in PBS | |||
| Buffer B: 0.5% w/v SDS, Protease inhibitor cocktail 2% v/v in PBS | |||
| Buffer C: N-glycosidase F reaction buffer: 200mM Na3PO4, pH 8.0, 4mM EDTA, 1.5% N-octylglucoside | |||
| NIH 3T3 stably expressing H2a 6 | |||
| Buffer D: 0.5% Triton X-100, 0.25% w/v sodium deoxycholate,0.5% w/v SDS, Protease inhibitor cocktail 2% v/v in PBS | |||
| Buffer E: 0.5% w/v SDS, 1% v/v 2-Mercapt–thanol, Protease inhibitor cocktail 2% v/v in PBS | |||
| 2-mercapt–thanol | Sigma-Aldrich | M3148 |
References
- Parodi, A. J., Behrens, N. H., Leloir, L. F., Carminatti, H. The role of polyprenol-bound saccharides as intermediates in glycoprotein synthesis in liver. Proc Natl Acad Sci U S A. 69 (11), 3268-3272 (1972).
- Avezov, E., Frenkel, Z., Ehrlich, M., Herscovics, A., Lederkremer, G. Z. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 19 (1), 216-225 (2008).
- Frenkel, Z., Gregory, W., Kornfeld, S., Lederkremer, G. Z. Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6-5GlcNAc2. J Biol Chem. 278 (36), 34119-34124 (2003).
- Hosokawa, N. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 278 (28), 26287-26294 (2003).
- Jakob, C. A., Burda, P., Roth, J., Aebi, M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 142 (5), 1223-1233 (1998).
- Tolchinsky, S., Yuk, M. H., Ayalon, M., Lodish, H. F., Lederkremer, G. Z. Membrane-bound versus secreted forms of human asialoglycoprotein receptor subunits. Role of a juxtamembrane pentapeptide. J Biol Chem. 271 (24), 14496-14503 (1996).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved