A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Purification of Hsp104, a Protein Disaggregase
In This Article
Summary
Here, we describe a protocol for the purification of highly active Hsp104, a hexameric AAA+ protein from yeast, which couples ATP hydrolysis to protein disaggregation. This scheme exploits a His6-tagged construct for affinity purification from E. coli followed by anion-exchange chromatography, His6-tag removal with TEV protease, and size-exclusion chromatography.
Abstract
Hsp104 is a hexameric AAA+ protein1 from yeast, which couples ATP hydrolysis to protein disaggregation2-10 (Fig. 1). This activity imparts two key selective advantages. First, renaturation of disordered aggregates by Hsp104 empowers yeast survival after various protein-misfolding stresses, including heat shock3,5,11,12. Second, remodeling of cross-beta amyloid fibrils by Hsp104 enables yeast to exploit myriad prions (infectious amyloids) as a reservoir of beneficial and heritable phenotypic variation13-22. Remarkably, Hsp104 directly remodels preamyloid oligomers and amyloid fibrils, including those comprised of the yeast prion proteins Sup35 and Ure223-30. This amyloid-remodeling functionality is a specialized facet of yeast Hsp104. The E. coli orthologue, ClpB, fails to remodel preamyloid oligomers or amyloid fibrils26,31,32.
Hsp104 orthologues are found in all kingdoms of life except, perplexingly, animals. Indeed, whether animal cells possess any enzymatic system that couples protein disaggregation to renaturation (rather than degradation) remains unknown33-35. Thus, we and others have proposed that Hsp104 might be developed as a therapeutic agent for various neurodegenerative diseases connected with the misfolding of specific proteins into toxic preamyloid oligomers and amyloid fibrils4,7,23,36-38. There are no treatments that directly target the aggregated species associated with these diseases. Yet, Hsp104 dissolves toxic oligomers and amyloid fibrils composed of alpha-synuclein, which are connected with Parkinson's Disease23 as well as amyloid forms of PrP39. Importantly, Hsp104 reduces protein aggregation and ameliorates neurodegeneration in rodent models of Parkinson's Disease23 and Huntington's disease38. Ideally, to optimize therapy and minimize side effects, Hsp104 would be engineered and potentiated to selectively remodel specific aggregates central to the disease in question4,7. However, the limited structural and mechanistic understanding of how Hsp104 disaggregates such a diverse repertoire of aggregated structures and unrelated proteins frustrates these endeavors30,40-42.
To understand the structure and mechanism of Hsp104, it is essential to study the pure protein and reconstitute its disaggregase activity with minimal components. Hsp104 is a 102kDa protein with a pI of ~5.3, which hexamerizes in the presence of ADP or ATP, or at high protein concentrations in the absence of nucleotide43-46. Here, we describe an optimized protocol for the purification of highly active, stable Hsp104 from E. coli. The use of E. coli allows simplified large-scale production and our method can be performed quickly and reliably for numerous Hsp104 variants. Our protocol increases Hsp104 purity and simplifies His6-tag removal compared to a previous purification method from E. coli47. Moreover, our protocol is more facile and convenient than two more recent protocols26,48.
Protocol
1. Expression of Hsp104
- The plasmid employed for purification in E. coli, pPROEX-HTb-Hsp104, contains the Hsp104 open-reading frame under the inducible control of the trc promoter26. The plasmid produces Hsp104 with an N-terminal His6-tag that can be removed by TEV protease cleavage. Transform pPROEX-HTb-Hsp104 into codon-optimized E. coli BL21-CodonPlus-RIL cells (Stratagene, Agilent Technologies) using a typical bacterial transformation procedure (e.g. according to manufacturer's instructions). It is important to use a codon-optimized E. coli strain because Hsp104 has an unusual codon bias.
- Inoculate a 100mL 2XYT culture (USB. Recipe: 16g/L casein peptone, 10g/L yeast extract, 5g/L NaCl, pH 7.0) supplemented with 100μg/mL ampicillin and 34μg/mL chloramphenicol with fresh transformants and grow overnight at 37°C with shaking at 200rpm.
- Inoculate 10mL of the overnight culture into 6 X 1L 2XYT with 100μg/mL ampicillin and 34μg/mL chloramphenicol in 2L flasks. Shake at 250rpm at 37°C until OD600 = 0.4-0.6. Stop shaking and reduce temperature to 15°C. Allow cells to equilibrate unagitated for 30min in the incubator until it reaches 15°C. Once 15°C is reached induce protein expression by adding IPTG to a final concentration of 1mM. Resume shaking at 250rpm overnight (12-16 hours).
2. Cell Harvest and Lysis
- Harvest cells by centrifugation (in a precooled rotor) at 4,000rpm for 20min at 4°C (we use a Sorvall RC 3BP+ centrifuge). Subsequent steps must be performed immediately because Hsp104 activity is diminished when E. coli cells are frozen. Moreover, all subsequent steps should be performed on ice or at 4°C.
- Resuspend cell pellets in prechilled (on ice) 10mL lysis buffer (40mM HEPES-KOH pH 7.4, 500mM KCl, 20mM MgCl2, 2.5% (w/v) glycerol, 20mM imidazole, 5μM pepstatin A, complete protease inhibitor cocktail (1 EDTA-free tablet/50mL), and 2mM β-mercaptoethanol).
- Lyse cells with a French press (Emulsiflex) homogenizer that uses a heat exchanger immersed in ice water. Before use, ensure that all cell clumps have been solubilized. After equilibrating homogenizer with lysis buffer, two passes through the cylinder with a pressure of 15,000-18,000psi is sufficient for full lysis. If a French press is not available, incubation with lysozyme followed by sonication can be used (see discussion). Save a small sample of lysed cells for SDS-PAGE analysis (1μL) (Lysate in Fig. 2).
- Remove cell debris by centrifugation at 16,000rpm for 20min at 4°C (we use a Sorvall RC5C+ centifuge). Retain supernatant for next step and save a small sample (1μL) of supernatant for SDS-PAGE analysis (Ni Load in Fig. 2).
3. Hsp104 Purification
- Mix supernatant from step 2.4 with 12mL (2mL Ni-Sepharose beads per 1L of cells) of 50% slurry of lysis buffer equilibrated Ni-Sepharose beads (GE).
- Rotate sample slowly for 3 hours at 4°C such that Ni-Sepharose remains evenly suspended and frothing is minimized. Shorter incubation times are possible, but will decrease yield. At 4°C in the presence of protease inhibitors, little degradation occurs, and a 3 hour Ni-Sepharose incubation time does not decrease activity of the final product. Collect Ni-Sepharose by centrifugation for 2 min at 4°C at 2,000rpm (Eppendorf 5810R centrifuge). Save a small sample of supernatant for SDS-PAGE analysis (1μL) and discard the rest (Ni Flow Through (FT) in Fig. 2).
- Wash the retrieved Ni-Sepharose with 25 column volumes of wash buffer (40mM HEPES-KOH pH 7.4, 150mM KCl, 20mM MgCl2, 2.5% (w/v) glycerol, 20mM imidazole, 2mM β-mercaptoethanol), 5 column volumes of wash buffer with 1M KCl to remove contaminants bound electrostatically to Hsp104, and 25 column volumes of wash buffer. Collect Ni-Sepharose after each wash by centrifugation for 2 min at 4°C at 2,000rpm (Eppendorf 5810R centrifuge). After each wash and centrifugation cycle, remove buffer by aspiration.
- To elute Hsp104, mix Ni-Sepharose with 1mL elution buffer (40mM HEPES-KOH pH 7.4, 150mM KCl, 20mM MgCl2, 2.5% (w/v) glycerol, 350mM imidazole, 2mM β-mercaptoethanol) per 1mL Ni-Sepharose and rotate at 4°C for 20 minutes. The high imidazole concentration disrupts the Ni bead-His6 interaction. Remove Ni-Sepharose with empty 1.2 mL spin columns (Bio-Rad) by centrifugation for 2 min at 4°C at 2,000rpm (Eppendorf 5810R centrifuge). Save 1μL of eluate for SDS-PAGE analysis (Ni eluate in Fig. 2). For every liter of cells, ˜15mg of Hsp104 is obtained at this step.
- Buffer exchange eluate into Buffer Q (20mM Tris-HCl pH 8, 0.5mM EDTA, 5mM MgCl2, 50mM NaCl) using MWCO 30,000 Amicon Ultra (Millipore) 15mL centrifugal concentrator units. First, concentrate protein to ˜1.5mL then add 14.5mL of Buffer Q to dilute protein 10-fold. Repeat 3 times for buffer exchange. The increased pH and low salt ensures that Hsp104 has a high negative charge.
- Purify Hsp104 via anion-exchange chromatography using a Buffer Q equilibrated Resource Q 6mL column (GE). Before injection, protein is filtered through a low protein binding Millex GP PES Membrane 0.22μm syringe filter (Millipore). We employ a flow rate of 1mL/min. Wash away weakly binding proteins with 1 column volume of 20% Buffer Q+ (20mM Tris-HCl pH 8, 0.5mM EDTA, 5mM MgCl2, 1M NaCl). Elute Hsp104 with linear gradient (20%-50% Buffer Q+) over 5 column volumes (Fig. 3). Collect 1ml fractions. Hsp104 and most variants typically elute at ˜400mM NaCl (31mS/cm). Save samples of peak fractions (1μL) for SDS-PAGE analysis (Fig. 3, inset).
- To remove His6-tag, exchange protein using the Amicon centrifugal concentrator as describe above (see step 3.5) into cleavage buffer (20mM HEPES-KOH pH 7.4, 140mM KCl, and 10mM MgCl2). Use proTEV protease (Promega) or AcTEV protease (Invitrogen) according to manufacturer's instructions. For Hsp104, we have found that higher ratios of TEV protease: Hsp104 are required for full cleavage. We use a ratio of 1 protease unit per 12μg Hsp104. Cleavage should be performed at 30°C for 2-4 hours, followed by a 16 hour incubation at 4°C. Final Hsp104 concentration should be between 20-75μM monomer. Deplete any remaining His6-tagged Hsp104 and TEV protease with Ni-Sepharose (GE) by adding Ni-Sepharose in excess to the amount of Hsp104 in the cleavage reaction (assume beads can bind 15mg of protein per mL of packed resin). Remove beads with empty spin columns (Bio-Rad) by centrifuging for 2min at 2,000rpm at 4°C. Collect samples before and after cleavage (1μL) for SDS-PAGE analysis to ensure cleavage, and the removal of uncleaved protein (Fig. 4).
- Exchange into size exclusion buffer (20mM HEPES-KOH pH 7.4, 140mM KCl, 10mM MgCl2, 0.1mM EDTA, 1mM DTT) using MWCO 30,000 Amicon Ultra (Millipore) as described in step 3.5.
- Further purify Hsp104 via size-exclusion chromatography. Use size exclusion buffer equilibrated Superose 6 or Superdex 200 column (both from GE) (Fig. 5). Before injection, protein is filtered through a low protein binding Millex GP PES Membrane 0.22μm syringe filter (Millipore). For less than 10mg of protein, the 10/300 size columns (24mL) can be used and 0.5ml fractions are collected. For samples greater than 10mg, the 12/60 size column (120mL) should be used and 1 ml fractions are collected. Refer to manufacturer's instructions for flow-rates and sample volumes to be loaded. After purification, Hsp104 fractions are pooled as shown in Fig. 5 and concentrated in Amicon Centrifugal Concentrator Units (Millipore) [Editor's Note: Video voice over incorrectly states 0.22μM (micromolar) syringe filter, when it should state 0.22μm (micrometer) syringe filter at timecode 04:38.]. Hsp104 is stored as described below. Due to loss of material in separating Hsp104 degradation products and contaminants the final yield of purified full-length Hsp104 is ˜1-3mg per liter of starting culture.
4. Hsp104 Disaggregase Activity
- After purification, it is good practice to assess Hsp104 activity in a disaggregation assay. Typically, we employ a luciferase reactivation assay2 (Fig. 6). In this assay, Hsp104 in conjunction with the Hsp70 chaperone system (Hsp70 and Hsp40) disaggregates urea-denatured firefly luciferase aggregates2. Soluble luciferase catalyzes the oxidation of luciferin to oxyluciferin, a reaction that releases light. By monitoring luminescence, the relative reactivation, and therefore disaggregation, of luciferase can be determined. Hsp104 must be able to synergistically collaborate with the Hsp70 co-chaperone system. The amount of recovered luminescence depends on the exact Hsp70:Hsp40 pair utilized. We routinely employ Hsp72 and Hsp40 (Assay Designs). At a minimum, active Hsp104 should produce a 5-fold increase in reactivation of luciferase when compared to reactivation by Hsp72:Hsp40 alone (Fig. 6). Hsp104 preparations that fail to reach this level of activity are discarded.
5. Hsp104 Storage
- For short-term storage, Hsp104 can be kept at 4°C in size-exclusion buffer. However, activity will decline after 2-3 days at 4°C and sharply after 1 week at 4°C. For disaggregation assays we recommend that Hsp104 should be used as soon as possible after purification. Ideally, Hsp104 should be used immediately for amyloid disaggregation assays.
- If protein must be stored long-term, Hsp104 is exchanged into storage buffer (20mM HEPES-KOH pH 7.4, 140mM KCl, 10mM MgCl2, 0.1mM EDTA, 1mM DTT, 10% glycerol (w/v)). 100μl aliquots are snap frozen in liquid nitrogen and stored at -80°C. Freeze-thaw cycles drastically reduce Hsp104 activity and should be avoided. We recommend slowly thawing Hsp104 on ice. Amyloid-disaggregase activity will decline sharply after 1 month at -80°C.
6. Representative Results and Figures:
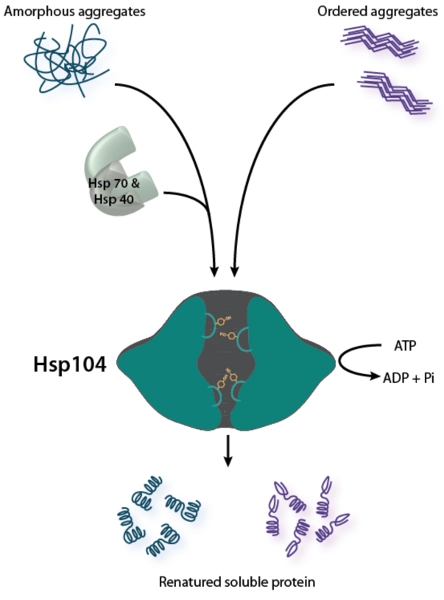
Figure 1. Hsp104 is a Bifunctional disaggregase. Disaggregation of disordered aggregates (shown on left) requires the cooperation of the Hsp70 chaperone system (Hsp70 and Hsp40)2. Hsp104 remodels ordered amyloid aggregates (shown on right) without the aid of Hsp70 and Hsp40 in vitro, but Hsp70 and Hsp40 can improve Hsp104 activity against amyloid26,28. For both types of aggregated structures, Hsp104 couples ATP hydrolysis to substrate translocation through its central channel to promote disaggregation. Tyrosine-bearing pore loops engage and shuttle substrate through the central channel49-52.

Figure 2. SDS-PAGE analysis of Ni-sepharose affinity purification step. Lysate, Ni Load, Ni FT and Ni eluate samples were fractionated by SDS-PAGE using a 4-20% Tris-HCl 1.0mm Criterion gel (Bio-Rad) and Coomassie stained. Note that not all the Hsp104 is able to bind to the Ni-sepharose. Broad Range molecular weight markers (Bio-Rad) are shown (left lane).
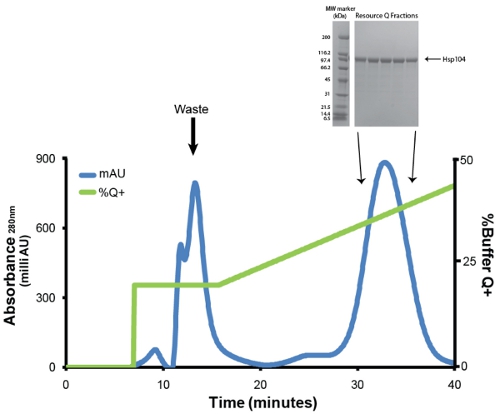
Figure 3. Resource Q purification of Ni-sepharose eluate. Blue trace represents absorbance at 280nm and green trace represents % Buffer Q+ (maximum at 50%). The first peak, which elutes at 20% Q+ contains impurities, degradation products and improperly folded Hsp104. The second and major peak contains properly folded and active Hsp104. Flow rate was 1ml/min. Gradient from 20-50% Q+ is 30 minutes or 5 column volumes. Inset: peak fractions are resolved by SDS-PAGE analysis using a 4-20% Tris-HCl 1.0mm Criterion gel (Bio-Rad) and Coomassie stained. Broad Range molecular weight markers (Bio-Rad) are shown (left).

Figure 4. SDS-PAGE analysis of proTEV protease cleavage step. His6-Hsp104 from Resource Q purification was treated with proTEV protease for 4 hours at 30°C and then 16 hours at 4°C. Samples were the fractionated by SDS-PAGE using a 4-20% Tris-HCl 1.0mm Criterion gel (Bio-Rad) and Coomassie stained. Note that proTEV cleaved Hsp104 migrates more rapidly. TEV protease and uncleaved Hsp104 have been depleted with Ni-Sepharose as described in Step 3.7. Broad Range molecular weight markers (Bio-Rad) are shown (left).
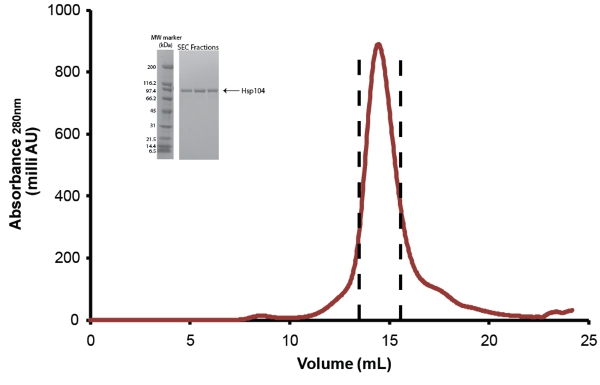
Figure 5. Size-exclusion purification of Hsp104. Cleaved Hsp104 was further purified via a Superose 6 gel filtration column (10/300, GE). Flow rate = 0.4ml/min. The peak between the dashed lines represents pooled fractions. Inset: peak fractions are resolved by SDS-PAGE analysis using a 4-20% Tris-HCl 1.0mm Criterion gel (Bio-Rad) and Coomassie stained. Broad Range molecular weight markers (Bio-Rad) are shown (left).
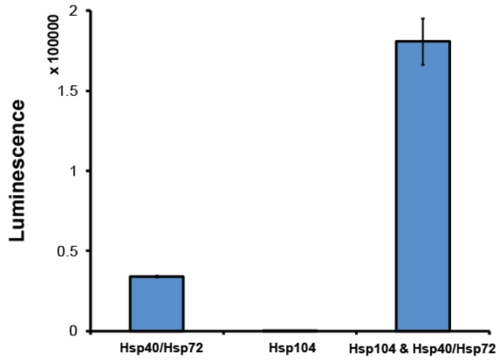
Figure 6. Luciferase reactivation assay. Denatured firefly luciferase aggregates (50nM) were incubated with either Hsp72 and Hsp40 (1μM) (both from Assay Designs), Hsp104 (6μM monomer) or Hsp104, Hsp72 and Hsp40 for 90min at 25°C. Denatured luciferase is only fully reactivated in the presence of both Hsp104 and Hsp40/Hsp72. Recovered luminescence is measured on an Infinite M1000 plate reader (Tecan). Values represent means±SEM (n=3).
Discussion
Timeline: For maximal Hsp104 activity we recommend that the entire purification scheme be completed as rapidly as possible. However, the number of purification steps makes a demanding schedule that may not always be practical. If the purification steps are carried out as quickly as possible, the time from the end of overnight expression through to the 2-4 hours of incubation at 30°C with TEV protease is approximately 9-11 hours. One potential place to pause is following the TEV cleavage step. If absolutely nec...
Disclosures
No conflicts of interest declared.
Acknowledgements
This work was supported by a grant from the NIH (5T32GM008275-22) and an American Heart Association predoctoral fellowship (to E.A.S.); a Chemistry-Biology Interface Fellowship from the NIH (2T32GM071339-06A1) (to M.E.D.); and grants from the NIH (1DP2OD002177-01 and NS067354-0110), The Ellison Medical Foundation, and The Bill and Melinda Gates Foundation (to J.S.).
Materials
| Name | Company | Catalog Number | Comments |
| BL21-CodonPlus-RIL Competent Cells | Stratagene, Agilent Technologies | 230255 | |
| 2XYT broth | USB Corp., Affymetrix | 75864 | |
| Complete, mini, EDTA-free protease inhibitor tablets | Roche Group | 1836170 | |
| Pepstatin A | Sigma-Aldrich | P4265 | |
| Ni-Sepharose 6 Fast Flow | GE Healthcare | 17-5318-02 | |
| Amicon Ultra-15 centrifugal filter units (MWCO 30,000) | EMD Millipore | UFC903008 | |
| Resource Q - 6ml column | GE Healthcare | 17-1179-01 | |
| proTEV Protease | Promega Corp. | V6052 | |
| AcTEV Protease | Invitrogen | 12575015 | |
| Superose 6 10/300 GL | GE Healthcare | 17-5172-01 | |
| Hsp40 | Assay Designs | SPP-400 | |
| Hsp72 | Assay Designs | ADI-NSP-555 |
References
- Erzberger, J. P., Berger, J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 35, 93-114 (2006).
- Glover, J. R., Lindquist, S. Hsp104, Hsp70, and Hsp40: A Novel Chaperone System that Rescues Previously Aggregated Proteins. Cell. 94, 73-82 (1998).
- Parsell, D. A., Kowal, A. S., Singer, M. A., Lindquist, S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 372, 475-478 (1994).
- Vashist, S., Cushman, M., Shorter, J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 88, 1-13 (2010).
- Parsell, D. A., Sanchez, Y., Stitzel, J. D., Lindquist, S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 353, 270-273 (1991).
- Doyle, S. M., Wickner, S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 34, 40-48 (2009).
- Shorter, J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 16, 63-74 (2008).
- Glover, J. R., Lum, R. Remodeling of protein aggregates by Hsp104. Protein Pept Lett. 16, 587-597 (2009).
- Mogk, A., Haslberger, T., Tessarz, P., Bukau, B. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans. 36, 120-125 (2008).
- Grimminger-Marquardt, V., Lashuel, H. A. Structure and function of the molecular chaperone Hsp104 from yeast. Biopolymers. 93, 252-276 (2010).
- Sanchez, Y., Lindquist, S. L. HSP104 required for induced thermotolerance. Science. 248, 1112-1115 (1990).
- Sanchez, Y., Taulien, J., Borkovich, K. A., Lindquist, S. Hsp104 is required for tolerance to many forms of stress. Embo J. 11, 2357-2364 (1992).
- Alberti, S., Halfmann, R., King, O., Kapila, A., Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 137, 146-158 (2009).
- Halfmann, R., Alberti, S., Lindquist, S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 20, 125-1233 (2010).
- Shorter, J., Lindquist, S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 6, 435-450 (2005).
- True, H. L., Berlin, I., Lindquist, S. L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 431, 184-187 (2004).
- True, H. L., Lindquist, S. L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 407, 477-483 (2000).
- Tyedmers, J., Madariaga, M. L., Lindquist, S. Prion switching in response to environmental stress. PLoS Biol. 6, e294-e294 (2008).
- Chernoff, Y. O., Lindquist, S. L., Ono, B., Inge-Vechtomov, S. G., Liebman, S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 268, 880-884 (1995).
- Halfmann, R., Lindquist, S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 330, 629-632 (2010).
- Satpute-Krishnan, P., Langseth, S. X., Serio, T. R. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 5, e24-e24 (2007).
- Sweeny, E. A., Shorter, J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2, 135-140 (2008).
- Lo Bianco, C. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 118, 3087-3097 (2008).
- Narayanan, S., Walter, S., Reif, B. Yeast prion-protein, sup35, fibril formation proceeds by addition and substraction of oligomers. Chembiochem. 7, 757-765 (2006).
- Savistchenko, J., Krzewska, J., Fay, N., Melki, R. Molecular chaperones and the assembly of the prion Ure2p in vitro. J Biol Chem. 283, 15732-15739 (2008).
- Shorter, J., Lindquist, S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 304, 1793-1797 (2004).
- Shorter, J., Lindquist, S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 23, 425-438 (2006).
- Shorter, J., Lindquist, S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. Embo J. 27, 2712-2724 (2008).
- Doyle, S. M. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 14, 114-122 (2007).
- Wendler, P. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 131, 1366-1377 (2007).
- Hinault, M. P. Stable alpha-synuclein oligomers strongly inhibit chaperone activity of the Hsp70 system by weak interactions with J-domain co-chaperones. J Biol Chem. 285, 38173-38182 (2010).
- Tipton, K. A., Verges, K. J., Weissman, J. S. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 32, 584-591 (2008).
- Bieschke, J., Cohen, E., Murray, A., Dillin, A., Kelly, J. W. A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis. Protein Sci1. 8, 2231-2241 (2009).
- Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W., Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science. 313, 1604-1610 (2006).
- Cohen, E. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 139, 1157-1169 (2009).
- Cushman, M., Johnson, B. S., King, O. D., Gitler, A. D., Shorter, J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 23, 1191-11201 (2010).
- Carmichael, J. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease. Proc Natl Acad Sci. 97, 9701-9705 (2000).
- Vacher, C., Garcia-Oroz, L., Rubinsztein, D. C. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum Mol Genet. 14, 3425-3433 (2005).
- Liu, Y. H. Heat Shock Protein 104 Inhibited the Fibrillization of Prion Peptide 106-126 and Disassembled Prion peptide 106-126 Fibrils in vitro. Int J Biochem Cell Biol. , (2011).
- Wendler, P., Saibil, H. R. Cryo electron microscopy structures of Hsp100 proteins: crowbars in or out. Biochem Cell Biol. 88, 89-96 (2010).
- Wendler, P. Motor mechanism for protein threading through Hsp104. Mol Cell. 34, 81-92 (2009).
- Lee, S., Sielaff, B., Lee, J., Tsai, F. T. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci. 107, 8135-8140 (2010).
- Parsell, D. A., Kowal, A. S., Lindquist, S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J Biol Chem. 269, 4480-4487 (1994).
- Schirmer, E. C., Queitsch, C., Kowal, A. S., Parsell, D. A., Lindquist, S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J Biol Chem. 273, 15546-15552 (1998).
- Schirmer, E. C., Ware, D. M., Queitsch, C., Kowal, A. S., Lindquist, S. L. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc Natl Acad Sci. 98, 914-919 (2001).
- Hattendorf, D. A., Lindquist, S. L. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 21, 12-21 (2002).
- Schirmer, E. C., Lindquist, S. Purification and properties of Hsp104 from yeast. Methods Enzymol. 290, 430-444 (1998).
- Hattendorf, D. A., Lindquist, S. L. Analysis of the AAA sensor-2 motif in the C-terminal ATPase domain of Hsp104 with a site-specific fluorescent probe of nucleotide binding. Proc Natl Acad Sci. 99, 2732-2737 (2002).
- Lum, R., Niggemann, M., Glover, J. R. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J Biol Chem. 283, 30139-30150 (2008).
- Lum, R., Tkach, J. M., Vierling, E., Glover, J. R. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 279, 29139-29146 (2004).
- Tessarz, P., Mogk, A., Bukau, B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 68, 87-97 (2008).
- Weibezahn, J. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 119, 653-665 (2004).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved