A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
High-throughput Measurement of Plasma Membrane Resealing Efficiency in Mammalian Cells
In This Article
Summary
Here we describe a high-throughput fluorescence-based assay that measures the plasma membrane resealing efficiency through fluorometric and imaging analyses in living cells. This assay can be used for screening drugs or target genes that regulate plasma membrane resealing in mammalian cells.
Abstract
In their physiological environment, mammalian cells are often subjected to mechanical and biochemical stresses that result in plasma membrane damage. In response to these damages, complex molecular machineries rapidly reseal the plasma membrane to restore its barrier function and maintain cell survival. Despite 60 years of research in this field, we still lack a thorough understanding of the cell resealing machinery. With the goal of identifying cellular components that control plasma membrane resealing or drugs that can improve resealing, we have developed a fluorescence-based high-throughput assay that measures the plasma membrane resealing efficiency in mammalian cells cultured in microplates. As a model system for plasma membrane damage, cells are exposed to the bacterial pore-forming toxin listeriolysin O (LLO), which forms large 30-50 nm diameter proteinaceous pores in cholesterol-containing membranes. The use of a temperature-controlled multi-mode microplate reader allows for rapid and sensitive spectrofluorometric measurements in combination with brightfield and fluorescence microscopy imaging of living cells. Kinetic analysis of the fluorescence intensity emitted by a membrane impermeant nucleic acid-binding fluorochrome reflects the extent of membrane wounding and resealing at the cell population level, allowing for the calculation of the cell resealing efficiency. Fluorescence microscopy imaging allows for the enumeration of cells, which constitutively express a fluorescent chimera of the nuclear protein histone 2B, in each well of the microplate to account for potential variations in their number and allows for eventual identification of distinct cell populations. This high-throughput assay is a powerful tool expected to expand our understanding of membrane repair mechanisms via screening for host genes or exogenously added compounds that control plasma membrane resealing.
Introduction
Mammalian cells are subject to mechanical, osmotic, and biochemical stress, resulting in the loss of plasma membrane integrity. Without rapid and efficient resealing, damaged cells would quickly succumb to programmed or necrotic death. Since the 1960s, efforts to understand the plasma membrane resealing process have been motivated by the devastating consequences associated with its dysfunctions. Indeed, diseases such as Limb-Girdle Muscular Dystrophy, diabetes, and Chediak-Higashi Syndrome have been linked to deficient plasma membrane repair due to mutations in the gene encoding dysferlin, production of advanced glycation end products, and defects in the lysosomal trafficking regulator CHS1, respectively1,2,3,4,5,6. However, to date, our understanding of membrane resealing is still limited7. Initial studies have demonstrated that membrane resealing is initiated by the influx of extracellular Ca2+ through the damaged plasma membrane8,9,10. Since then, several non-mutually exclusive Ca2+-dependent mechanisms have been proposed to reseal cells. The patch hypothesis proposes that in proximity to the wound, intracellular vesicles fuse with each other and the damaged plasma membrane to act as a patch11,12,13,14. A second model proposes that calcium-dependent exocytosis of lysosomes at the wound site releases the lysosomal enzyme acid sphingomyelinase, which converts sphingomyelin to ceramide in the outer leaflet of the plasma membrane. This sudden change in lipid composition results in ceramide-driven endocytosis of the damaged region15,16,17. Lastly, the third proposed mechanism involves a role for the endosomal sorting complex required for transport (ESCRT) to promote the formation of outward-facing vesicles that bud off from the plasma membrane18. Only a limited set of proteins was identified in these models, and their machinery must be further elucidated.
Here we describe a high-throughput assay that measures the plasma membrane resealing efficiency in adherent mammalian cells subjected to damage mediated by recombinant listeriolysin O (LLO)19. LLO is a pore-forming toxin (PFT) secreted by the facultative intracellular pathogen Listeria monocytogenes20,21,22 and belongs to the MACPF/CDC (membrane attack complex, perforin, and cholesterol-dependent cytolysin) superfamily. MACPF are mammalian pore-forming proteins involved in immune defenses, whereas CDCs are bacterial toxins mainly produced by Gram-positive pathogens that damage host cells to promote their pathogenic lifestyles23. CDCs are synthesized as water-soluble monomers or dimers that bind to cholesterol present in the plasma membrane and oligomerize into a prepore complex of up to 50 subunits. The prepore complex then rearranges to insert β-strands across the lipid bilayer, forming a β-barrel pore that spans 30-50 nm in diameter24,25,26,27. These large pores permit fluxes of ions and small cellular components in and out of the cell; though, some studies have proposed that pores of smaller sizes are also formed28,29,30. Among the CDCs, LLO displays unique properties including irreversible pH- and temperature-dependent aggregation, which is conducive to high-throughput analyses31,32. LLO can be added to the cell culture medium at 4 ˚C, a temperature permissive to its binding to cells, but not to the formation of the pore complex. Initiation of pore formation can then be synchronized by raising the temperature to 37 ˚C, allowing for the efficient diffusion of toxin molecules in the plane of the membrane to form oligomers and for the conformational remodeling involved in pore generation. Therefore, following the switch in temperature, the kinetic of cell damage will depend on the amount of toxin bound to the plasma membrane. Importantly, soluble LLO (not bound to the plasma membrane) rapidly and irreversibly aggregates when the temperature reaches 37 ˚C, which alleviates the need to wash away unbound toxin molecules and limits the extent of membrane damage over time. Lastly, because LLO binds to cholesterol and forms pores in cholesterol-rich membranes, this assay is amenable to a wide range of mammalian cells. It is important to keep in mind that LLO affects host cell signaling mainly via pore formation, with a few exceptions in which pore-independent cell signaling may occur33,34,35,36,37,38,39. Therefore, it cannot be excluded that LLO signaling activities may influence the process of membrane repair.
This assay directly assesses the extent of cell wounding by measuring the incorporation of a cell impermeant fluorochrome (e.g., propidium iodide) that passively enters wounded cells and becomes highly fluorescent once it associates with nucleic acids. Hence, the fluorochrome can be maintained in the cell culture medium throughout the experiment, allowing real-time analyses of cell wounding. The fluorescence intensity of the nucleic acid-binding dye will increase with the concentration of toxin and, for a given concentration of toxin, will increase over time until all pores are formed, and cells are fully repaired or until saturation is reached. The influx of extracellular Ca2+ through membrane pores is a sine qua non event for resealing. Therefore, the resealing efficiency can be indirectly evidenced by comparing cell wounding in culture medium containing Ca2+ (repair permissive condition) to wounding in a Ca2+-free medium (repair restrictive condition). Because the fluorescence intensity of the nucleic acid-binding dye is directly proportional to the cell concentration in each well, it is important to seed cells at the same concentration in all wells. It is also important to enumerate cells in each well before and after the assay to ensure that cell detachment does not occur, as floating, aggregated cells can obscure fluorescence readings which may complicate data interpretation. To enumerate cells, cells expressing nuclear-localized histone 2B-GFP (H2B-GFP) were used in this assay. Temperature-controlled, multi-mode, microplate readers combine rapid, high-throughput measurements (using a 96 or 384-well plate format) of fluorescence intensities with microscopy imaging of living cells at 37 °C. The latter can be used to enumerate cell number and observe the eventual formation of distinct cell populations.
Ultimately, this assay provides users the ability to expand their knowledge of the complexity of membrane repair mechanisms by screening for host molecules or exogenously added compounds that may control membrane repair. The following protocol describes the experimental steps to measure the resealing efficiency of cells exposed to LLO and evaluate the effects of a given drug or cellular treatment on resealing efficiency.
Protocol
1. Preparation
- Cell Plating
Note: Human cervical epithelial cells, HeLa and HeLa expressing Histone 2B-GFP (H2B-GFP), were used in this protocol, but this assay can be adapted to other mammalian cells19.- Detach adherent cells from a 75 cm2 cell culture flask by washing the cells with 2 mL of Trypsin-EDTA 0.25%. Replace the used trypsin with 2 mL of fresh trypsin-EDTA 0.25%.
- Incubate the cells at 37 ˚C for 5 min until the cells have rounded and detached from the flask.
- Resuspend the cells in 8 mL of growth medium (DMEM containing 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin).
- Determine the cell concentration using a hemocytometer and 10 µL of cell suspension.
- Dilute the cells in growth medium to a concentration of 2.5 x 105 cells/mL.
- Pour the cell suspension into a sterile pipette basin and thoroughly mix the suspension using a 10 mL serological pipette.
- Using a 12-multichannel micropipette and 200 µL tips, distribute HeLa cells (2.5 x 104 cells/100 µL/well) in triplicate (or quadruplicate) in a 96-well flat, clear bottom, black polystyrene tissue culture-treated plate.
Note: A plating arrangement is presented as an example in Figure 1. - Culture the cells for 24 h in a humidified cell culture incubator at 37 ˚C and 5% CO2.
- Stock Solution Preparation
- Prepare 1 L of a 10x stock of buffer M (used to prepare M1 and M2) by adding 95 g of Hanks Balanced Salt Solution, 0.476 g of MgCl2 (5 mM), and 23.83 g of HEPES (100 mM) to 900 mL of water. Adjust the pH to 7.4 and raise the volume to 1 L. Filter sterilize.
- Prepare 50 mL of a 50x (1.25 M) stock of glucose by adding 11.26 g of D-(+)-Glucose to a total of 50 mL of water. Filter sterilize the solution.
- Prepare 50 mL of a 100x (120 mM) stock of calcium by adding 0.666 g of CaCl2 to a total of 50 mL of water. Filter sterilize the solution.
- Prepare 50 mL of a 10x (50 mM) stock of ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’,tetraacetic acid (EGTA) by adding 0.951 g of EGTA to 40 mL of water. Increase the pH to 8 using NaOH to dissolve the EGTA, then raise the volume to 50 mL. Filter sterilize the solution.
- For a single 96-well plate, prepare 50 mL of Medium 1 (M1, contains Ca2+), 50 mL of Medium 2 (M2, Ca2+-free), and 15 mL of Medium 2 supplemented with EGTA, accordingly:
- For M1, add 5 mL of 10x Buffer M, 0.5 mL of 100x CaCl2, and 1 mL of 50x glucose to 43.5 mL of water.
- For M2, add 5 mL of 10x Buffer M and 1 mL of 50x glucose to 44 mL of water.
- For M2/EGTA, add 1.5 mL of 10x Buffer M and 1.5 mL of 10x EGTA to 12 mL of water.
Note: All solutions containing propidium iodide (PI) should be prepared directly prior to adding to the cells.
- Plate Reader/Imaging Cytometer Settings
Note: Use a multi-mode plate reader equipped with two detection units: a spectrofluorometer and an imaging cytometer. Limit the fluorescence exposure to avoid photobleaching the fluorophores.- Pre-warm the plate reader to 37 °C before performing the assay.
- Set up the parameters for the kinetic assay accordingly within the Settings mode:
- Choose Monochromator, FL (fluorescence), and Kinetic for the optical configuration, read modes, and read type, respectively.
- Under Wavelength Settings, select a 9 and 15 nm excitation and emission bandpass, respectively. For assays using propidium iodide (PI), set the excitation and emission wavelengths to 535 and 617 nm, respectively.
- Under Plate Type, select 96 Wells for the plate format and a pre-set plate configuration corresponding to a black-wall clear bottom plate.
- Under Read Area, highlight the wells that will be analyzed throughout the kinetic.
- Under PMT and Optics, preset the flashes per read to 6 and check the box for Read from Bottom.
- Under Timing, insert 00:30:00 in the Total Run Time box for a 30 min kinetic assay, and insert 00:05:00 for the Interval.
Note: For each time point and one wavelength, the reading time of a full 96-well plate is 30 s. - Confirm the specified settings in the Settings Information to the right and select OK. Press Read to initiate the kinetic run.
- Set up the imaging parameters accordingly within the Settings mode:
- Choose Minimax, Imaging, and Endpoint for the optical configuration, read modes, and read type, respectively.
- Under Wavelengths, select transmitted light, and either or both the fluorescence boxes corresponding to excitation and emission wavelengths of 456/541 nm (GFP) and 625/713 nm (PI).
- Use the same options for the Plate Type and Read Area as defined in steps 1.3.2.3 and 1.3.2.4.
- Under Well Area Setting, select the number of sites within a well to be imaged.
Note: 12 sites correspond to a full-well image. - Under the Image Acquisition Settings, select the exposure times for transmitted light, 541 (GFP), and 713 (PI). For GFP, image the entire well with an exposure time of 20 ms/image. For transmitted light (TL) and PI fluorescence, acquire a single image of the center of each well with exposure times of 8 and 20 ms, respectively.
- Confirm the specified settings in the Settings Information to the right and select OK. The acquisition time for imaging the entire surface of each well (12 images/well) of a 96-well plate and for one wavelength is ~15 min. Press Read to initiate imaging.
Note: The acquisition time of a single image/well of a 96-well plate requires ~2.5 min/plate for one wavelength. The parameters described above correspond to the specific equipment in our laboratory. Spectrofluorometric measurements: A xenon flash lamp displaying 1.0 nm increment excitation wavelengths (250-850 nm) with an adjustable 9 or 15 nm bandpass, a photomultiplier tube detector with a > 6 log dynamic range and an adjustable 15 or 25 nm emission bandpass. Imaging cytometer: An illumination light source capable of white light, 460 nm and 625 nm excitation wavelengths with a 20 nm bandpass, emission filters centered at 541 nm (108 nm bandpass) and 713 nm (123 nm bandpass), respectively, and a 4X objective coupled to a 1.25 megapixel 12-bit charge-coupled device camera.
2. Assay
Note: At the time of the assay, cells must be 70-90% confluent. During the wash steps, the medium should be removed from and applied to the side-wall of the well (not directly above the cells). Maintain the temperature of LLO at < 4 ˚C to prevent its aggregation until step 3.1.5.
- Prepare a stock of 30 µM PI in M1 and a stock of 30 µM PI in M2 pre-warmed at 37 ˚C.
- Gently wash the cells in plate 1 using a 12-multichannel micropipette and 200 µL tips, as follows:
- For repair-permissive conditions, remove the growth medium and wash the cells twice with 200 µL/well M1 pre-warmed at 37 ˚C. Replace the medium with 100 µL/well of warm M1 containing 30 µM PI.
- For repair restrictive-conditions, remove the growth medium and wash the cells once with 200 µL/well warm M2 containing 5 mM EGTA to chelate Ca2+, followed by one wash with 200 µL/well M2. Replace the medium with 100 µL/well warm M2 containing 30 µM PI.
- After the growth medium has been washed and replaced with medium containing propidium iodide, directly move to step 2.1.3.
- Image plate 1 under transmitted light, GFP, and PI as detailed under 1.3.3 (pre-kinetic). This step takes 15-20 min.
- During the 15 min period in step 2.1.3, prepare plate 2 using a 12-multichannel micropipette and 200 µL tips as follows:
- Place a 96-well round bottom polypropylene microplate on ice. Configure the plate using an experimental design corresponding to plate 1 (Figure 1).
- For repair-permissive conditions, add 100 µL/well of ice-cold M1 containing 60 µM PI, followed by the addition of 100 µL/well of ice-cold M1 containing 4x LLO or not for the control.
- For repair-restrictive conditions, add 100 µL/well of ice-cold M2 containing 60 µM PI, followed by the addition of 100 µL/well of ice-cold M2 containing 4x LLO or not for the control.
- After imaging plate 1 (step 2.1.3), immediately place it on ice, using aluminum foil to separate the plate from direct contact with ice. Allow plate 1 to cool down for 5 min.
- Using a 12-multichannel micropipette and 200 µL tips, transfer 100 µL from each well into plate 2 (step 2.1.4) to the corresponding wells in the plate 1. To properly distribute the toxin in the media of plate 1, insert the tips below the meniscus and gently eject the volume without introducing bubbles.
Note: Do not pipette up and down, as this may inadvertently detach the cells. - Leave the plate for an additional 1 min to allow the toxin to bind to the cells and immediately transfer plate 1 to the plate reader for the kinetic assay using the spectrofluorometer mode (step 1.3.2).
- At the end of the kinetic assay, immediately image plate 1 (post-kinetic) using step 1.3.3.
3. Analysis: Cell Enumeration
- Determine the cell count based on the nuclear fluorescence using the microplate cell enumeration software.
- Within Settings, select Re-analysis, and under the category section within the Image Analysis Settings select Discreet Object Analysis using 541 as the wavelength for finding objects.
- Within the Find Objects option, using the Draw on Images finding method, select Nuclei under the settings tab, and press Apply.
- Press OK and Read to initiate the cell counting algorithm.
- Alternatively, if no such tool is available, use an image analysis software such as ImageJ to enumerate cells.
- In ImageJ, open the image file as a stack.
- Convert the stack to 8-bit greyscale images by clicking Image in the menu bar, hover over Type, and select 8-bit.
- Subtract the background: Click Image in the menu bar, hover over Adjust, and select Brightness/Contrast. Adjust the minimum value to remove the background noise and select Apply.
- Threshold to create binary images: Click Image in the menu bar, hover over Adjust, and select Threshold. Select Dark background, adjust the minimum and maximum threshold values, and click Apply.
- In the case of overlapping nuclei, a Watershed tool can be used to segment nuclei. Click Process in the menu, hover over Binary and select Watershed.
Note: This will automatically separate connected nuclei. - Analyze the masked images by applying user-specified criteria (size and circularity) to refine the identification of nuclei and exclude cell debris.
- Click on Analyze in the menu and then Analyze particles. Set the desired size (pixel^2) and circularity (a value of 1 is a perfect circle) ranges that are sufficient to include individual cells/nuclei.
- In the Show dropdown box, select the option(s) desired, check Summarize, and click OK to obtain cell counts.
4. Analysis: Kinetic Curves
- Transfer the kinetic data from the plate reader software to an analytical data software.
- For each experimental condition, average the fluorescence intensities of the replicates at each timepoint, along with the corresponding standard deviation and standard error of the mean for each experimental condition.
- For each experimental condition, trace the corresponding kinetic curve: PI intensity (y-axis) versus time (x-axis).
- To calculate the resealing efficiency of a given treatment condition, calculate the area under the curve (AUC) of the +LLO in M1 (AUC(M1)) and +LLO in M2 (AUC(M2)). Use the approach suggested below to assess the efficiency (E) of resealing:
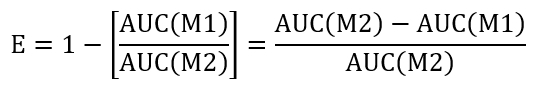
- Perform a comparison between control and test treatment by determining the efficiency ratio (REff) indicated below:
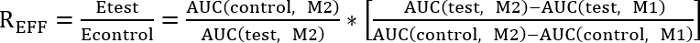
REFF = 1, test treatment has no effect on repair
REFF < 1, test treatment inhibits repair
REFF > 1, test treatment improves repair - Calculate the area under the curve using the following equation:
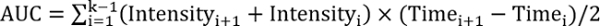 , where k is the total number of follow-ups.
, where k is the total number of follow-ups.
Results
Cell counting accuracy: HeLa cells are frequently used as a model mammalian cell line to explore membrane repair mechanisms. When assessing membrane repair at the cell population level, it is important to plate cells at the same concentration in all wells for proper data interpretation. It is also important to verify at the time of the assay that cell numbers are equivalent across wells. HeLa cells that constitutively express histone 2B fused to GFP (H2B-GFP) were introduced in this assay...
Discussion
This assay measures the efficiency of membrane resealing at the cell population level with high-throughput capacity. It can be used to screen for cellular components or drug libraries that could affect membrane repair. The described assay used a 96-well plate format, but it can be adapted to 384-well plates for higher throughput. An advantage of this assay is its ability to obtain fluorescence measurements of adherent living cells in real time without the need for excessive cell processing such as cell detachment, fixati...
Disclosures
The authors have nothing to disclose.
Acknowledgements
We acknowledge Dr. Jesse Kwiek (The Ohio State University) for kindly allowing us to use his multi-mode detection platform for some preliminary experiments. Research reported in this article was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number RO1AI107250 to Stephanie Seveau. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| Name | Company | Catalog Number | Comments |
| SpectraMax i3x Multi-Mode Microplate Reader | Molecular Devices | i3x | |
| MiniMax 300 Imaging cytometer | Molecular Devices | 5024062 | |
| TO-PRO-3 | ThermoFisher Scientific | T3605 | |
| Propidium Iodide | ThermoFisher Scientific | P3566 | |
| HeLa | ATCC | CCL2 | |
| HeLa H2B-GFP | Millipore | SCC117 | |
| Trypsin-EDTA 0.25% | ThermoFisher Scientific | 25200056 | |
| 96-well Corning flat bottom black polystyrene tissue culture treated plate | Corning | 3603 | |
| Hanks' balanced Salts | Sigma-Aldrich | H4891 | |
| EGTA | ISC BioExpress | 0732-100G | |
| HEPES | Fisher Scientific | BP310-500 | |
| D-(+)-Glucose, HybriMax | Sigma-Aldrich | G5146-1KG |
References
- Demonbreun, A. R., McNally, E. M. Plasma Membrane Repair in Health and Disease. Current Topics in Membranes. 77, 67-96 (2016).
- Howard, A. C., McNeil, A. K., McNeil, P. L. Promotion of plasma membrane repair by vitamin E. Nature Communications. 2, 597 (2011).
- Howard, A. C., et al. A novel cellular defect in diabetes: membrane repair failure. Diabetes. 60 (11), 3034-3043 (2011).
- Lozano, M. L., et al. Towards the targeted management of Chediak-Higashi syndrome. Orphanet Journal of Rare Diseases. 9, 132 (2014).
- Vainzof, M., et al. Dysferlin protein analysis in limb-girdle muscular dystrophies. Journal of Molecular Neuroscience. 17 (1), 71-80 (2001).
- Huynh, C., et al. Defective lysosomal exocytosis and plasma membrane repair in Chediak-Higashi/beige cells. Proceeding of the National Academy of Sciences of the United States of America. 101 (48), 16795-16800 (2004).
- Cooper, S. T., McNeil, P. L. Membrane Repair: Mechanisms and Pathophysiology. Physiological Reviews. 95 (4), 1205-1240 (2015).
- Steinhardt, R. A., Bi, G., Alderton, J. M. J. M. Cell membrane resealing by a vesicular mechanism similar to neurotransmitter release. Science. 263 (5145), 390-393 (1994).
- De Mello, W. C. Membrane sealing in frog skeletal-muscle fibers. Proceedings of the National Academy of Sciences of the United States of America. 70 (4), 982-984 (1973).
- Fishman, H. M., Tewari, K. P., Stein, P. G. Injury-induced vesiculation and membrane redistribution in squid giant axon. Biochimica et Biophysica Acta. 1023 (3), 421-435 (1990).
- Davenport, N. R., Bement, W. M. Cell repair: Revisiting the patch hypothesis. Communicative & Integrative Biology. 9 (6), 1253643 (2016).
- McNeil, P. L., et al. Patching plasma membrane disruptions with cytoplasmic membrane. Journal of Cell Science. 113 (11), 1891-1902 (2000).
- Terasaki, M., Miyake, K., McNeil, P. L. Large plasma membrane disruptions are rapidly resealed by Ca2+-dependent vesicle-vesicle fusion events. Journal of Cell Biology. 139 (1), 63-74 (1997).
- Bi, G. Q., Alderton, J. M., Steinhardt, R. A. Calcium-regulated exocytosis is required for cell membrane resealing. Journal of Cell Biology. 131 (6 Pt. 2), 1747-1758 (1995).
- Tam, C., et al. Exocytosis of acid sphingomyelinase by wounded cells promotes endocytosis and plasma membrane repair. Journal of Cell Biology. 189 (6), 1027-1038 (2010).
- Rodriguez, A., et al. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. Journal of Cell Biology. 137 (1), 93-104 (1997).
- Reddy, A., Caler, E. V., Andrews, N. W. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 106 (2), 157-169 (2001).
- Jimenez, A. J., et al. ESCRT machinery is required for plasma membrane repair. Science. 343 (6174), 1247136 (2014).
- Pathak-Sharma, S., et al. High-Throughput Microplate-Based Assay to Monitor Plasma Membrane Wounding and Repair. Frontiers in Cellular and Infection Microbiology. 7, 305 (2017).
- Hamon, M. A., et al. Listeriolysin O: the Swiss army knife of Listeria. Trends in Microbiology. 20 (8), 360-368 (2012).
- Seveau, S. Multifaceted activity of listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Subcellular Biochemistry. 80, 161-195 (2014).
- Osborne, S. E., Brumell, J. H. Listeriolysin O: from bazooka to Swiss army knife. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 372 (1726), (2017).
- Lukoyanova, N., Hoogenboom, B. W., Saibil, H. R. The membrane attack complex, perforin and cholesterol-dependent cytolysin superfamily of pore-forming proteins. Journal of Cell Science. 129 (11), 2125-2133 (2016).
- Tweten, R. K. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infection and Immunity. 73 (10), 6199-6209 (2005).
- Koster, S., et al. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nature Communications. 5, 3690 (2014).
- Duncan, J. L., Schlegel, R. Effect of streptolysin O on erythrocyte membranes, liposomes, and lipid dispersions. A protein-cholesterol interaction. Journal of Cell Biology. 67 (1), 160-174 (1975).
- Morgan, P. J., et al. Subunit organisation and symmetry of pore-forming, oligomeric pneumolysin. FEBS Letters. 371 (1), 77-80 (1995).
- Leung, C., et al. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. eLife. 3, (2014).
- Marchioretto, M., et al. What planar lipid membranes tell us about the pore-forming activity of cholesterol-dependent cytolysins. Biophysical Chemistry. 182, 64-70 (2013).
- Palmer, M., et al. Assembly mechanism of the oligomeric streptolysin O pore: the early membrane lesion is lined by a free edge of the lipid membrane and is extended gradually during oligomerization. European Molecular Biology Organization Journal. 17 (6), 1598-1605 (1998).
- Bavdek, A., et al. pH dependence of listeriolysin O aggregation and pore-forming ability. Federation of European Biochemical Society Journal. 279 (1), 126-141 (2012).
- Schuerch, D. W., Wilson-Kubalek, E. M., Tweten, R. K. Molecular basis of listeriolysin O pH dependence. Proceeding of the National Academy of Sciences of the United States of America. 102 (35), 12537-12542 (2005).
- Cassidy, S. K., O'Riordan, M. X. More than a pore: the cellular response to cholesterol-dependent cytolysins. Toxins (Basel). 5 (4), 618-636 (2013).
- Lam, J., et al. Host cell perforation by listeriolysin O (LLO) activates a Ca(2+)-dependent cPKC/Rac1/Arp2/3 signaling pathway that promotes L. monocytogenes internalization independently of membrane resealing. Molecular Biology of the Cell. , (2017).
- Gekara, N. O., Weiss, S. Lipid rafts clustering and signalling by listeriolysin O. Biochemical Society Transactions. 32 (Pt 5), 712-714 (2004).
- Magassa, N., Chandrasekaran, S., Caparon, M. G. Streptococcus pyogenes cytolysin-mediated translocation does not require pore formation by streptolysin O. European Molecular Biology Organization Reports. 11 (5), 400-405 (2010).
- Baba, H., et al. Induction of gamma interferon and nitric oxide by truncated pneumolysin that lacks pore-forming activity. Infection and Immunity. 70 (1), 107-113 (2002).
- Carrero, J. A., Vivanco-Cid, H., Unanue, E. R. Listeriolysin o is strongly immunogenic independently of its cytotoxic activity. Public Library of Science One. 7 (3), e32310 (2012).
- Coconnier, M. H., et al. Listeriolysin O-induced stimulation of mucin exocytosis in polarized intestinal mucin-secreting cells: evidence for toxin recognition of membrane-associated lipids and subsequent toxin internalization through caveolae. Cell Microbiology. 2 (6), 487-504 (2000).
- Suzuki, T., et al. DNA staining for fluorescence and laser confocal microscopy. Journal of Histochemistry and Cytochemistry. 45 (1), 49-53 (1997).
- Bink, K., et al. TO-PRO-3 is an optimal fluorescent dye for nuclear counterstaining in dual-colour FISH on paraffin sections. Histochemistry and Cell Biology. 115 (4), 293-299 (2001).
- Zhang, J. H., Chung, T. D., Oldenburg, K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening. 4 (2), 67-73 (1999).
- Birmingham, A., et al. Statistical methods for analysis of high-throughput RNA interference screens. Nature Methods. 6 (8), 569-575 (2009).
- Zhang, X. D. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 89 (4), 552-561 (2007).
- Zhang, X. D. A new method with flexible and balanced control of false negatives and false positives for hit selection in RNA interference high-throughput screening assays. Journal of Biomolecular Screening. 12 (5), 645-655 (2007).
- Idone, V., et al. Repair of injured plasma membrane by rapid Ca2+-dependent endocytosis. Journal of Cell Biology. 180 (5), 905-914 (2008).
- Davenport, N. R., et al. Membrane dynamics during cellular wound repair. Molecular Biology of the Cell. 27 (14), 2272-2285 (2016).
- Defour, A., Sreetama, S. C., Jaiswal, J. K. Imaging cell membrane injury and subcellular processes involved in repair. Journal of Visualized Experiments. (85), (2014).
- Lee, J. J. A., et al. Cell Membrane Repair Assay Using a Two-photon Laser Microscope. Journal of Visualized Experiments. (131), (2018).
- Weisleder, N., et al. Visualization of MG53-mediated cell membrane repair using in vivo and in vitro systems. Journal of Visualized Experiments. (52), (2011).
- Corrotte, M., et al. Toxin pores endocytosed during plasma membrane repair traffic into the lumen of MVBs for degradation. Traffic. 13 (3), 483-494 (2012).
- Kuismanen, E., Saraste, J. Low temperature-induced transport blocks as tools to manipulate membrane traffic. Methods in Cell Biology. 32, 257-274 (1989).
- Togo, T., et al. The mechanism of facilitated cell membrane resealing. Journal of Cell Science. 112, 719-731 (1999).
- Johnson, S. A., et al. Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochimica et Biophysica Acta. 1798 (7), 1427-1435 (2010).
- Lam, J. G. T., et al. Host cell perforation by listeriolysin O (LLO) activates a Ca(2+)-dependent cPKC/Rac1/Arp2/3 signaling pathway that promotes Listeria monocytogenes internalization independently of membrane resealing. Molecular Biology of the Cell. 29 (3), 270-284 (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved