A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Methylation Specific Multiplex Droplet PCR using Polymer Droplet Generator Device for Hematological Diagnostics
In This Article
Summary
Epigenetic markers are used for white blood cell (WBC) subtyping through the quantification of DNA methylation patterns. This protocol presents a multiplex droplet polymerase chain reaction (mdPCR) method using a thermoplastic elastomer (TPE)-based microfluidic device for droplet generation allowing for precise and multiplex methylation-specific target quantification of WBC differential counts.
Abstract
A multiplexed droplet PCR (mdPCR) workflow and detailed protocol for determining epigenetic-based white blood cell (WBC) differential count is described, along with a thermoplastic elastomer (TPE) microfluidic droplet generation device. Epigenetic markers are used for WBC subtyping which is of important prognostic value in different diseases. This is achieved through the quantification of DNA methylation patterns of specific CG-rich regions in the genome (CpG loci). In this paper, bisulfite-treated DNA from peripheral blood mononuclear cells (PBMCs) is encapsulated in droplets with mdPCR reagents including primers and hydrolysis fluorescent probes specific for CpG loci that correlate with WBC sub-populations. The multiplex approach allows for the interrogation of many CpG loci without the need for separate mdPCR reactions, enabling more accurate parametric determination of WBC sub-populations using epigenetic analysis of methylation sites. This precise quantification can be extended to different applications and highlights the benefits for clinical diagnosis and subsequent prognosis.
Introduction
Analysis of white blood cells (WBCs) composition is among the most frequently requested laboratory tests in hematological diagnostics. Differential leukocyte count serves as an indicator for a spectrum of diseases including infection, inflammation, anemia, and leukemia, and is under investigation as an early prognostic biomarker for several other conditions as well. Gold standard in WBC subtyping involves immunostaining and/or flow cytometry both of which require costly, instability-prone fluorescent antibodies and are often highly dependent on operator proficiency in sample preparation. Moreover, this method is applicable to fresh blood samples only, such that the samples cannot be frozen for shipment or later analysis.
Epigenetic markers have recently emerged as powerful analytical tools for the study of phenotypic variations. Subsequently, human leukocyte populations have been shown to have cell-lineage DNA methylation patterns that allow for the precise characterization of WBC subsets. Subtyping based on epigenetic markers provides a promising alternative that does not depend on fresh blood sample collection or expensive antibodies and can be exploited as a biomarker for disease onset and susceptibility1,2,3,4,5.
Genome-wide studies have been performed for extensive mapping of methylated specific CG-rich regions in the genome (CpG islands) in leukocyte subtypes to identify candidate epigenetic markers specific to leukocyte subtypes. PCR protocols have been developed because of this reason for methylated gene regions, e.g., CD3Z and FOXP3, corresponding to CD3+ T-Cells and CD4+ CD25+ Regulatory T-Cells (T-Regs), respectively. Wiencke et al. have demonstrated the utility of duplex droplet PCR for epigenetic subtyping of T-Cells, yielding results that highly correlate with flow activated cell sorting (FACS) analysis6. This quantitative genetic analysis method relies on partitioning the template nucleic acid molecules and PCR reagents into thousands of discrete, volumetrically defined, sub-nanoliter sized droplets containing zero, one or more target nucleic acid copies, using water-in-oil emulsions enabled by microfluidics7,8. The PCR amplification is performed within each individual droplet and the endpoint fluorescence intensity of each droplet is measured, allowing absolute quantification of targets present in the sample. Droplet PCR has been established to be more precise, accurate, and technically simpler than standard qPCR, making it a more favorable DNA methylation-based method for clinical evaluation of T-Cells. Although a rapidly emerging subtyping methodology, multiplexed epigenetic analysis to probe for various methylated CpG regions simultaneously is lacking. This is necessary for routine leukocyte differential counts.
Herein, a thermoplastic elastomer (TPE) droplet microfluidic device is presented and employed for methylation-specific multiplex droplet PCR (mdPCR). The technology has been used to delineate specific leukocyte subtypes, CD3+ T-Cells and CD4+ CD25+ T-Regs, based on cell-lineage DNA methylation patterns, i.e., epigenetic variation of CD3Z and FOXP3 CpG regions, respectively. A detailed protocol for DNA extraction, bisulfite conversion and mdPCR is described in concert with a fabrication method for a TPE droplet generation device. Representative results of the method are compared to those of immunofluorescence staining highlighting the utility of the proposed approach.
Protocol
All the experiments performed in this study involving human samples were approved by the NRC’s Ethics Board and were done according to NRC’s policies governing human subjects that follow applicable research guidelines and are compliant with the laws in Québec, Canada.
1. Cell preparation
- Thaw the frozen human peripheral blood mononuclear cells (PBMCs) immediately by placing the cryovial in a water bath at 37 °C for 5 min.
- Invert the cryovial twice to gently resuspend the cells and using a 1 mL pipette transfer the cell suspension into a 15 mL conical tube.
- Add 10 mL of pre-warmed (37 °C) growth medium supplemented with 10% fetal bovine serum (RPMI-1640 + 10% FBS) to the 15 mL tube containing the PBMCs.
- Centrifuge the cell suspension in a swinging bucket centrifuge at room temperature at a speed of 330 x g for 10 min with rapid acceleration and the brake on high.
- Once the spin is over, carefully decant the supernatant. Resuspend the cell pellet in 3 mL of phosphate-buffered saline (PBS) pH 7.2, containing 2 mM ethylenediaminetetraacetic acid (EDTA) by tapping the side of the tubes.
- Mix the cells by inverting the tube with the cap tightly closed.
- Prepare two 1.5 mL microtubes and using a pipette aliquot 1.5 mL of the cell suspension in each tube, one of which is used for subsequent immunofluorescence staining and one for the DNA extraction.
2. Immunofluorescence staining and imaging protocol
- Resuspend cells (from 1.7) in 200 μL of PBS buffer containing 0.1% sodium azide and 2% FBS and adjust the final concentration of the cell suspension to a maximum of 2 x 107 cells/mL.
- Divide the cell suspension by pipetting 100 μL volume in two separate 1.5 mL microtubes.
- Add 20 μL volume of anti-Hu CD3/CD4 conjugated with Fluorescein isothiocyanate (FITC) and Phycoerythrin (PE) to one tube and anti-Hu CD4/CD25 conjugated with FITC and PE (see Table of Materials) to the second tube, respectively.
- Add 1 drop of blue fluorescent live cell stain (see Table of Materials) to each tube.
- Incubate at room temperature using a tube rotator for 2 h. Protect from light.
- Centrifuge the cell suspension at room temperature at 330 x g for 10 min with rapid acceleration and the brake on high.
- Decant the supernatant and carefully resuspend the cell pellet by tapping the tube. Add 1 mL of PBS, pH 7.2 containing 2 mM EDTA. Ensuring that the cap is tightly closed, mix the cells by inverting the tube 2x.
- Repeat steps 2.6 and 2.7 for three times.
- Resuspend cells in 20 μL of PBS pH 7.2 containing 2 mM EDTA.
- Pipette 10 μL drop of the cell suspension onto a borosilicate microscope slide and wait for 2 min for cells to slowly sediment to the bottom of the drop.
- Carefully place a glass cover slip on top of the microscope slide and place the slide on the stage of an inverted microscope.
- Record images of the cells using a 10x objective and an EMCCD camera connected to the microscope for each of the fluorophores for both cell suspension samples.
- Manually count fluorescently labeled cells (see Supplementary Information for raw data).
- Take the ratio of anti-Hu CD3 and anti-Hu CD4/CD25 labeled-cells to DAPI-stained cells to obtain the proportions of CD3+ T-Cells and CD4+ CD25+ T-Regs to total leukocyte.
3. DNA extraction and bisulfite conversion
- DNA extraction
NOTE: Extract DNA from PBMCs prepared in section 1 using a magnetic DNA purification kit (see Table of Materials) following procedures provided by the manufacturer.- In a 1.5 mL tube suspend cells in 100 µL of PBS and add 20 µL of Proteinase K and 400 µL of Lysis/Binding buffer. Mix by pipetting up-down 10x, then perform incubation at room temperature for 5 min.
- Capture the DNA-bead complex by placing the tube on a magnetic rack for 1-2 min, then carefully remove and discard the supernatant.
- Remove the tube containing the DNA-bead complex from magnetic rack and resuspend the beads in 600 µL of Wash Buffer #1 to wash away any non-specific binding.
- Place the tube again on the magnetic rack and carefully remove and discard the supernatant.
- Repeat steps 3.1.3 and 3.1.4 with 600 µL of Wash Buffer #2.
- Leave the tube open to air-dry for 1 min.
- Remove the tube from the magnetic rack and elute the DNA by dispensing 100 µL of Elution Buffer and pipetting the DNA/bead complex up and down 20x.
- Place the tube containing the eluted DNA again on the magnetic rack and incubate for 1-2 min to separate the magnetic beads from the eluted DNA.
- Transfer the eluted purified DNA solution to a new clean tube.
- Assess the concentration of the purified DNA sample by measuring the absorbance at 260 nm using a spectrophotometer.
- Bisulfite conversion
NOTE: Perform the bisulfite conversion on purified DNA using a methylation kit (see Table of Materials) following procedures provided by the manufacturer.- To 20 µL of DNA sample (200-500 ng) in a PCR tube add 130 µL of Conversion Reagent. Mix well and spin down briefly.
- Transfer the PCR tube to a thermal cycler and perform the cycling protocol as follows: 98 °C for 8 min; 54 °C for 60 min and hold at 4 °C.
- Add 600 µL of the Binding buffer to an ion chromatography (IC) column placed into a collection tube.
- Add the DNA sample to the IC column containing the binding buffer and mix by inverting the tube several times. Centrifuge for 30 s at full speed. Discard the collected flow-through.
- To the column now add 100 µL of Wash Buffer. Perform centrifugation as described in step 3.2.4 and discard the flow-through.
- Add 200 µl of Desulfonation Buffer and perform incubation at room temperature for 15-20 min. Centrifuge and discard the flow-through as described in steps above.
- Add 200 µL of Wash Buffer to the column. Centrifuge at full speed for 30 s and discard the flow-through.
- Repeat step 3.2.7.
- Transfer the column to a new 1.5 mL collection tube and add 100 µL of PCR-grade water on the membrane of the column. Centrifuge at full speed for 1 min to elute the DNA.
- Assess the concentration of bisulfite-converted DNA sample by measuring the absorbance at 260 nm using a spectrophotometer.
NOTE: Use a value of 40 µg/mL for absorbance at 260 nm = 1.0. - Store DNA at -20 °C for short term storage or at -70 °C for long term storage.
4. Droplet generation device fabrication
NOTE: A microfluidic device used for droplet generation (CAD file provided in the Supplementary Information) was fabricated in a clean room (class 1,000) environment in thermoplastic elastomer (see Table of Materials) using hot embossing generated by the following protocol.
- SU-8 mold fabrication
- Prepare an SU-8 mold on a 6″ silicon wafer using standard photolithography as detailed below.
- Clean a 6” silicon wafer using oxygen plasma at 500 W for 10 s.
- Spin-coat SU-8 resist onto the silicon wafer at 900 rpm for 40 s to achieve a total film thickness of 100 µm.
- Place the wafer on a hot plate and pre-bake for 15 min at 65 °C, followed by 2 h at 95 °C.
- Expose to UV light at 365 nm (Hg i-line) through a high-definition transparency photomask using an exposure dose of 1,000 mJ/cm2.
- Place the wafer on a hot plate and post-bake for 15 min at 65 °C, followed by 40 min at 95 °C.
- Develop by immersing in propylene glycol monomethyl ether acetate (PGMEA) for 5 min.
- Rinse with PGMEA and isopropanol and dry with a stream of nitrogen gas.
- Place the wafer on a hot plate and hard-bake for 2 h at 135 °C.
- Silanize the wafer in the vacuum desiccator containing a drop of silanizing agent (tricholoro perfluorooctyl silane) placed on an adjacent glass microscope slide for 2 h.
NOTE: This is done to make the silanes form a monolayer on the surface of the SU-8 master. Tricholoro perfluorooctyl silane should be always handled in the fume hood and kept away from water sources.
- Polydimethylsiloxane (PDMS) replica
- Prepare liquid prepolymers of PDMS (see Table of Materials) at a 10:1 ratio w/w of elastomer base to curing agent in a plastic cup.
- Place the cup in a planetary centrifugal vacuum mixer to mix and degas the PDMS mixture.
- Pour PDMS mixture onto the mold placed in the custom metal holder that prevents resin leakage and cure at 65 °C for 2 h.
- Using tweezers, carefully peel off the PDMS mold from the SU-8 master.
- Epoxy mold
NOTE: An epoxy mold was fabricated from the SU-8/silicon master using an intermediate replication process with PDMS.- Prepare the epoxy resin (see Table of Materials) using a 100/83 w/w ratio of resin/hardener.
- Degas the mixture under reduced pressure using a vacuum drying oven for 30 min.
- Pour the resin over the PDMS replica and cure at 80 °C for 12 h.
- Remove the cured epoxy mold from the PDMS replica, place on a hot plate and hard-bake for 2 h at 120 °C.
- TPE device
- Extrude pellets of TPE (see Table of Materials) at 165 °C in 2.0 mm thick and 7” wide sheets of several meters in length and store them as a roll for future use.
- Cut the TPE sheet from the roll using scissors into a 7” square.
- Place the TPE sheet between the epoxy mold and a non-patterned silanized silicon wafer (see step 4.1.10 for wafer silanization procedure).
- Perform hot-embossing at a temperature of 125 °C, an applied force of 10 kN, and a pressure of 10–2 mbar for 10 min.
- Demold carefully at room temperature using methanol spray to separate the embossed TPE from the silicon wafer and the epoxy mold.
- Cut another 7” square sheet of TPE and place it between two non-patterned silanized silicon wafers.
- Perform hot-embossing at a temperature of 140 °C, an applied force of 10 kN, and a pressure of 10–2 mbar for 10 min to form a planar surface for closing the channels and sealing the device.
- Demold carefully at room temperature using methanol spray to separate the embossed TPE from the two silicon wafers.
- Cut each of the embossed TPE sheets to device size using a doctor blade.
- Punch the access holes for the inlet and outlet channels in the structured device using a 1 mm biopsy punch needle with a plunger.
- Enclose the channels by placing a planar TPE device in direct contact with the channels at room temperature.
- Optionally, place in an oven at 70 °C for 2 h to promote device bonding.
- Fit the access holes with a disposable fluidic tubing (I.D. 0.25 mm, O.D. 0.8 mm). Seal the joints using an epoxy glue to ensure leak-proof manipulation.
5. Droplet generation and PCR
NOTE: Table 1 outlines information on the forward and reverse primers along with the double-quenched hydrolysis probes for C-LESS, CD3Z and Foxp3 genes, which are required for the multiplex amplification of demethylated gene targets.
- Prepare the master mix as described in Table 2.
- Thaw all components of the master mix except for the enzyme mix. Mix the master mix thoroughly by pipetting up-down and spin down briefly.
- Add the appropriate volume (1 μL) of bisulfite converted DNA (from section 3.2) to master mix in a PCR tube. Mix the reaction by pipetting up-down and spin down briefly.
- Connect disposable fluidic tubing (I.D. 0.25 mm, O.D. 1.6 mm) to two precision glass syringes (250 μL volume) using PEEK fittings.
- Prefill one precision glass syringe with 250 μL of carrier oil containing 5% fluoro-surfactant.
- Prefill another precision glass syringe with 50 µL of carrier oil before loading 100 µL of the PCR mix to ensure dispensing of the entire sample volume during emulsification.
- Set-up a droplet microfluidic device on a stage of an upright light microscope equipped with a high-speed camera to observe and record droplet formation in real-time.
- Place the prefilled syringes onto the programmable syringe pump and using PEEK union with fittings (see Table of Materials), connect the tubing of the syringes to the tubing of the respective inlet channels of the droplet microfluidic device.
- Place the tubing from the outlet of the droplet generator inside a 0.5 mL PCR tube.
- Adjust the flow rate of the syringe pump to 2 µL/min and allow for the droplet size to stabilize before collecting the resulting emulsion.
- Collect the emulsion and transfer 75 μL to a 0.2 mL PCR tube for thermal cycling.
- Ensure that the oil content in the PCR tube closely matches the volume of the dispersed phase in order to prevent coalescence of the droplets during thermal cycling.
- Place the 0.2 mL PCR tube in thermal cycler and perform the cycling protocol as follows: preheating at 95 °C for 5 min, then 45 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 30 s.
- Use the remaining emulsion to fill a borosilicate capillary tube (100 μm depth) with a rectangular profile in order to image droplets and assess the droplet diameter.
- Place the borosilicate tube filled with the emulsion onto a microscope slide. Use an inverted microscope equipped with an EMCCD camera and 10x objective to record bright field images of the droplets.
- Measure the droplet diameter using an image analysis software as detailed below.
- Set the scale of known distance in microns corresponding to the number of pixels in the image by selecting the ‘Analyze’ button and then ‘Set Scale’.
- Convert the image to grayscale by selecting the ‘Image’ button and then ‘Type’. Adjust brightness and contrast if needed. Set a manual threshold to delineate and fill the circles by selecting the ‘Image’ button and then ‘Adjust Threshold’.
- Analyze the particles by selecting the ‘Analyze particles’ button and setting the circularity to 0.75 – 1.
- Obtain the resulting area and the diameter of the measured droplets which is automatically displayed in the software.
- Compute the mean droplet diameter and assuming a spherical droplet, estimate the partition volume, which will be used to calculate absolute target concentration.
- Ensure that the droplets are monodisperse by analyzing the coefficient of variation (CV) which is taken as the ratio of the standard deviations to the mean values (k = 1), for the droplet diameter (< 3%).
6. Fluorescence imaging and image analysis
- Fluorescence imaging
- After amplification, transfer the PCR emulsion into borosilicate capillary tube (50 μm depth) with a rectangular profile in order to arrange droplets into a close-packed monolayer for imaging.
- Fix the filled capillaries on a microscope slide and seal both sides using a UV acrylic adhesive. Apply a UV light source on the UV adhesive being careful not to illuminate the emulsion to avoid bleaching the sample.
- Load the glass slide on an inverted microscope equipped with an EMCCD camera and a 10x objective.
- Using microscope imaging software, select Acquire | Live - Fast to start real-time camera acquisition, observe the sample, and ensure that the width of the capillary is captured.
- Set the bright field lamp for diascopic illumination at 3.5 V.
- Set the broad-spectrum LED fluorescent lamp for episcopic illumination at 20% intensity.
- Adjust the capture setting for all wavelengths (Bright field, FAM, HEX and Cy5) manually by selecting the Calibration | Optical Configurations command in the imaging software. For each fluorophore, the corresponding fluorescence filter cube needs to be manually switched. A filter cube for the bright field imaging must be used to keep the same optical path.
- Adjust the exposure time manually prior to image acquisition using Acquire | Camera Settings command for each fluorophore as summarized in Table 3. Set the readout mode EM gain 17 MHz at 16-bit with a gain multiplier of 100 in the ‘Capture Settings’ menu of the software.
- In the LUT window of the software, set the LUTs scale such that the transmission signal is encompassed within the set range. In this experiment, the scale was set from 500 up to 12,000 approximately.
- Automate the capture using a multipoint acquisition program using both XY scan and multiple wavelength options in that specific order. Ensure that the stage moves to the initial position, capture all the different wavelengths, and then proceed to the next position.
- First, set-up the XY scan by customizing the XY scan profile in the XY scan menu of the software. In ‘Custom Multipoint Definition’, choose the large image definition box and set it to 40.0 x 1.0 mm approximately (the length of the emulsion filling). Use 1% overlap.
- Enable the ‘Use Focus Surface’ option and set up the focus surface curve by adjusting the focus plane at different points on the sample using the focus knob on the microscope.
- Second, setup the multiple wavelength scan by selecting the scan tab. Add each of the optical configurations created in step 6.1.7 for each wavelength.
- Click the option to close active shutter during stage movement and during filter change to avoid bleaching the sample and run the acquisition by pressing the ‘Start Run’ button. Export each acquisition frame into tiff files and split each channel into a separated file using split multiple files option and by applying saved LUTs settings. Use point name and channel name option to easily differentiate each image file.
- Image analysis
- Sort all acquired images by brightfield and fluorescence filters to upload to an open source image analysis software.
- Use image analysis software to create a pipeline in order to identify all droplets using brightfield images, then measure intensity of associated fluorescent droplets.
- To pipeline, upload brightfield and fluorescence tiff images then add modules ‘ColorToGray’, ‘IdentifyPrimaryObjects’, ‘MeasureObjectIntensity’, and ‘ExportToSpreadsheet’. Use brightfield droplet images to identify objects, then use objects as mask to measure intensity of fluorescent images.
- Run pipeline with selected tiff images to extract average fluorescence intensity of fluorescence droplet images. Conduct the experiment in triplicate, with each set consisting of ~5,000 droplets for analysis.
- Apply the ‘definetherain’ algorithm (http://definetherain.org.uk/) to identify the positive and negative droplet clusters. The positives should be within 3 standard deviations of the mean. This determines the threshold intensity of positive droplets to be used for counting.
- Again use the image analysis software to implement a new pipeline where the fluorescence threshold for each gene target is set as defined in the previous step.
- Upload brightfield and fluorescent images to pipeline. Add modules ‘ColorToGray’, ‘RescaleIntensity’, ‘Threshold’, ‘IdentifyPrimaryObjects’, ‘MeasureObjectSizeShape’, ‘FilterObjects’, and ‘ExportToSpreadsheet’. Create unique modules for brightfield images and each fluorescence filter for droplets.
- Rescale image intensity scale from 0 to 1 for each fluorescence image group. Then set the threshold to identify and count the objects above the established threshold. If necessary, add ‘ExpandOrShrinkObjects’ module to shrink objects to facilitate the counting and identification of droplets in brightfield.
- Only identify objects within selected size of 20-30 pixels and filter the counted objects to only retain those objects with a specific diameter (i.e., droplets in the 75 µm diameter range) and a round spherical eccentricity of 0.5 and below.
- Export the results to a table that lists the total droplet count from brightfield images, as well as droplet counts of all the fluorescence channels used; namely Cy5 for C-LESS gene, HEX for methylated CD3Z gene, and FAM for methylated FOXP3 gene (see Supplementary Information for raw experimental data obtained for the presented mdPCR assay).
- Calculate the ratio of negative droplets for each gene target and apply Poisson distribution to obtain the respective copies per droplet (CPD) using Equation 1:
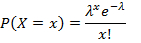
where x represents the number of droplets containing 0, 1, 2 or more molecules, and λ represents the CPD value. - Calculate, the absolute target concentration by taking the ratio of the CPD value and the droplet volume obtained in step 5.20 with Equation 2:
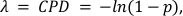
where the value (1-p) represents the fraction of negative droplets. - Calculate the percentage of CD3+ T-Cells and CD4+ CD25+ T-Regs by dividing the respective CPD values of methylated CD3Z and FOXP3 genes by the C-LESS – or total cell – CPD value (see Supplementary Information for CPD calculations from raw data).
- Compare these percent values to those obtained from immunofluorescence imaging using antibodies for CD3+ T-Cell and CD4+ CD25+ T-Reg counting.
Results
The TPE-based microfluidic droplet generator device was fabricated using the described protocol as shown in Figure 1. A transparency mask was used in photolithography to obtain silicon (Si) master. Soft lithography was performed to obtain an inverse PDMS replica of the Si master which was then used to fabricate the epoxy mold. Epoxy precursor was poured onto the PDMS and cured to crosslink and harden. This mold, representing the exact replica of the Si master was more resilient for subsequen...
Discussion
The presented experimental protocol and methods allow for in-house mdPCR using a fabricated TPE droplet generator, a thermal cycler, and fluorescence microscope. The fabricated device using soft TPE to TPE bonding affords hydrophobic surface properties that are uniform across all channel walls, such that the final device does not require any surface treatment for subsequent use as a droplet generator. This material has been routinely employed in point-of-care platforms that necessitate compatibility with high throughput ...
Disclosures
There are no conflicts to declare.
Acknowledgements
The authors acknowledge financial support from the National Research Council of Canada.
Materials
| Name | Company | Catalog Number | Comments |
| Bio-Rad, Mississauga, ON | TFI0201 | PCR tube | |
| RAN Biotechnologies, Beverly, MA | 008-FluoroSurfactant | Fluoro-surfactant | |
| Silicon Quest International, Santa Clara, CA | |||
| Oxford Instruments, Abingdon, UK | EMCCD camera | ||
| Thermo Fisher Scientific, Waltham, MA | MA5-16728 | ||
| Thermo Fisher Scientific, Waltham, MA | 22-8425-71 | ||
| CellProfiler | Used for fluorescence image analysis | ||
| Nikon, Japan | 10x objective | ||
| American Type Culture Collection (ATCC), Manassas, VA | PCS-800-011 | ||
| Ramé-Hart Instrument Co. (Netcong, NJ) | p/n 200-U1 | ||
| Fisher, Canada | |||
| Vitrocom, NJ, USA | 5015 and 5010 | Borosilicate capilary tube | |
| (http://definetherain.org.uk/) | |||
| Hamamatsu, Japan | LC-L1V5 | DEL UV light source | |
| Dolomite | 3200063 | Disposable fluidic tubing | |
| Dolomite | 3200302 | Disposable fluidic tubing | |
| IDT, Coralville, IA | |||
| Nikon, Melville, NY | Upright light microscope | ||
| Cytec Industries, Woodland Park, NJ | |||
| EV Group, Schärding, Austria | |||
| Zymo Research, Irvine, CA | D5030 | ||
| Photron, San Diego, CA | |||
| IDT, Coralville, IA | |||
| Gersteltec, Pully, Switzerland | SU-8 photoresist | ||
| Fineline Imaging, Colorado Springs, CO | |||
| Qiagen, Hilden, Germany | 203603 | ||
| Image J | Used to assess droplet diameter | ||
| Anachemia, Montreal, QC | |||
| Excelitas, MA, USA | Broad-spectrum LED fluorescent lamp | ||
| Galenvs Sciences Inc., Montreal, QC | DE1010 | ||
| Hexpol TPE, Åmål, Sweden | Thermoplastic elastomer (TPE) | ||
| Thermo Fisher Scientific, Waltham, MA | 13-400-518 | ||
| Nikon, Japan | Used for image acquisition | ||
| 3M, St Paul, MN | Carrier Oil | ||
| Thermo Fisher Scientific, Waltham, MA | R37605 | Blue fluorescent live cell stain (DAPI) | |
| IDEX Health & Science, Oak Harbor, WA | P-881 | PEEK fittings | |
| Sigma-Aldrich, Oakville, ON | 806552 | ||
| Dow Corning, Midland, MI | |||
| ThinkyUSA, CA, USA | ARV 310 | ||
| Ihc world, Maryland, USA | IW-125-0 | ||
| Zinsser NA, Northridge, CA | 2607808 | ||
| Cetoni GmbH, Korbussen, Germany | |||
| Sigma-Aldrich, Oakville, ON | 484431 | ||
| Bio-Rad, Mississauga, ON | 1861096 | ||
| Hitachi High-Technologies, Mississauga, ON | |||
| Nikon, Melville, NY | Inverted microscope | ||
| Nikon, Japan | |||
| Loctite | AA 352 |
References
- Teitell, M., Richardson, B. DNA methylation in the immune system. Clinical Immunology. 109 (1), 2-5 (2003).
- Suarez-Alvarez, B., Rodriguez, R. M., Fraga, M. F., López-Larrea, C. DNA methylation: A promising landscape for immune system-related diseases. Trends in Genetics. 28 (10), 506-514 (2012).
- Suárez-Álvarez, B., Raneros, A. B., Ortega, F., López-Larrea, C. Epigenetic modulation of the immune function: A potential target for tolerance. Epigenetics. 8 (7), 694-702 (2013).
- Kondilis-Mangum, H. D., Wade, P. A. Epigenetics and the adaptive immune response. Molecular Aspects of Medicine. 34 (4), 813-825 (2013).
- Zouali, M. . The Autoimmune Diseases. , (2014).
- Wiencke, J. K., et al. A comparison of DNA methylation specific droplet digital PCR (mdPCR) and real time qPCR with flow cytometry in characterizing human T cells in peripheral blood. Epigenetics. 9 (10), 1360-1365 (2014).
- Hindson, B. J., et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry. 83 (22), 8604-8610 (2011).
- Pinheiro, L. B., et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Analytical Chemistry. 84 (2), 1003-1011 (2012).
- Roy, E., Galas, J. C., Veres, T. Thermoplastic elastomers for microfluidics: Towards a high-throughput fabrication method of multilayered microfluidic devices. Lab on a Chip. 11 (18), 3193-3196 (2011).
- Roy, E., et al. From cellular lysis to microarray detection, an integrated thermoplastic elastomer (TPE) point of care Lab on a Disc. Lab on a Chip. 15 (2), 406-416 (2015).
- Malic, L., et al. Epigenetic subtyping of white blood cells using a thermoplastic elastomer-based microfluidic emulsification device for multiplexed, methylation-specific digital droplet PCR. Analyst. 44 (22), 6541-6553 (2019).
- Malic, L., et al. Polymer-based microfluidic chip for rapid and efficient immunomagnetic capture and release of Listeria monocytogenes. Lab Chip. 15 (20), 3994-4007 (2015).
- Malic, L., Morton, K., Clime, L., Veres, T. All-thermoplastic nanoplasmonic microfluidic device for transmission SPR biosensing. Lab Chip. 13 (5), 798-810 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved