A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Preparation of Tunable Extracellular Matrix Microenvironments to Evaluate Schwann Cell Phenotype Specification
In This Article
Summary
This methodology aims to illustrate the mechanisms by which extracellular matrix cues such as substrate stiffness, protein composition and cell morphology regulate Schwann cell (SC) phenotype.
Abstract
Traumatic peripheral nervous system (PNS) injuries currently lack suitable treatments to regain full functional recovery. Schwann cells (SCs), as the major glial cells of the PNS, play a vital role in promoting PNS regeneration by dedifferentiating into a regenerative cell phenotype following injury. However, the dedifferentiated state of SCs is challenging to maintain through the time-period needed for regeneration and is impacted by changes in the surrounding extracellular matrix (ECM). Therefore, determining the complex interplay between SCs and differing ECM to provide cues of regenerative potential of SCs is essential. To address this, a strategy was created where different ECM proteins were adsorbed onto a tunable polydimethylsiloxane (PDMS) substrate which provided a platform where stiffness and protein composition can be modulated. SCs were seeded onto the tunable substrates and critical cellular functions representing the dynamics of SC phenotype were measured. To illustrate the interplay between SC protein expression and cellular morphology, differing seeding densities of SCs in addition to individual microcontact printed cellular patterns were utilized and characterized by immunofluorescence staining and western blot. Results showed that cells with a smaller spreading area and higher extent of cellular elongation promoted higher levels of SC regenerative phenotypic markers. This methodology not only begins to unravel the significant relationship between the ECM and cellular function of SCs, but also provides guidelines for the future optimization of biomaterials in peripheral nerve repair.
Introduction
Peripheral nervous system (PNS) injuries remain a major clinical challenge in healthcare by compromising the quality of life for patients and creating a significant impact through a multitude of socioeconomic factors1,2. Schwann cells (SC), as the major glial cells in the PNS, provide necessary molecular and physical cues to induce PNS regeneration and aid in functional recoveries in short gap injuries. This is due to the remarkable ability of SCs to dedifferentiate into a “repair” cell phenotype from a myelinating or Remak phenotype3. The repair SC is a distinctive cell phenotype in several ways. Following injury, SCs increase their proliferation rate by re-entering the cell cycle and begin expression of several transcriptional factors to facilitate reinnervation. These factors, such as c-Jun and p75 NTR, are upregulated while myelinating SC markers, such as myelin basic protein (MBP), are downregulated4,5. In addition, SCs change morphology to become elongated and aligned with each other to form Büngner bands across the injury site6. This provides a physical guidance mechanism for the axons to extend to the correct distal target7. However, despite the ability that SCs possess to promote nerve regeneration in short gap injuries, the outcome of functional recovery remains poor in severe injuries. This is due in part to a loss of extracellular matrix (ECM) guidance cues, as well as the inability of SCs to maintain the regenerative phenotype over long periods of time8.
The nerve regeneration and recovery process are intimately tied to the state of the basal lamina following injury. The basal lamina is a layer of ECM around the nerve that facilitates guidance and provides ECM-bound cues for axons and SCs in cases where it remains intact following injury9. The state of the ECM and its ability to deliver matrix bound cues to cells is vitally important and has been previously explored in a variety of different contexts10,11,12,13,14. For example, it has been shown that the stiffness of the ECM can guide cell functions such as proliferation and differentiation11,15,16. Composition of the ECM can also lead to a distinct cellular response and regulate cell behaviors such as migration and differentiation through intracellular signaling pathways17,18. Furthermore, cell morphology, including spreading area and cellular elongation, play a major role in regulating the function and can be governed by ECM-bound cues19,20. Many previous studies have focused on stem cells differentiating into defined lineages, yet SCs possess a similar ability to alter phenotype from a homeostatic, adult SC within a healthy nerve, to a repair SC capable of secreting proteins and growth factors while remodeling the ECM following nerve injury5,21. Therefore, it is especially crucial to identify mechanisms underlying the relationship between the innate SC regenerative capacity and ECM bound cues for the insight to ultimately harness this capacity for nerve regeneration.
To address this, we have developed a detailed methodology to produce a cell culture substrate where mechanical stiffness and ligand type can be easily tuned in physiologically relevant ranges. Polydimethyl siloxane (PDMS) was chosen as a substrate due to its highly tunable mechanics as compared to polyacrylamide gel, where the maximum Young’s modulus is around 12 kPa contrasted to PDMS at around 1000 kPa22,23,24. This is beneficial to the work at hand, as recent studies have shown the Young’s modulus of a rabbit sciatic nerve can exceed 50 kPa during development, thereby suggesting that the range of stiffness of nerves within the PNS is wider than previously examined. Different proteins are capable of adsorption onto PDMS substrates to analyze the combinatorial regulation of mechanics and ligands on SC behavior. This allows for the investigation of multiple microenvironmental cues present in the PNS regeneration process and comparison of a high degree of tunability to the work focusing solely on stiffness of the substrate25. Further, these engineered cell culture substrates are compatible with a multitude of quantitative analysis methods such as immunohistochemistry, western blot, and quantitative polymerase chain reaction (q-PCR).
This engineered cell culture platform is highly suitable for analyzing mechanistic pathways due to the high level of individual tunability of each ECM-bound signal. In addition, popular methods for cell micropatterning, including microcontact printing, can be achieved on the substrates to allow for controlled cellular adhesion to analyze cell shape in relation to other ECM bound cues24. This is critical because line patterned substrates, which promote elongation in cell populations, provide a tool to mimic and study elongated and regenerative SCs within Büngner bands during nerve regeneration. Further, cellular morphology is a potent regulator of multiple cell functions and can potentially introduce confounding experimental results if not controlled26,27. Significant attention is now being provided to the mechanisms governing the SC regenerative phenotype as regulated by ECM cues28,29,30. This is essential to provide insight into the design of biomaterials that can be applied as nerve guidance conduits for aid in PNS nerve regeneration. These detailed protocols can ultimately be applied as a potential tool to decipher the mechanisms of SC and other cell type function as regulated by ECM bound cues.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Tunable cell culture substrate preparation and characterization
- Substrate preparation
- Mix the PDMS base elastomer and curing agents using a pipette tip vigorously at a ratio between 10:1 and 60:1 until bubbles are homogeneously dispersed within the mixture. Remove bubbles using vacuum desiccation until bubbles are dissipated.
NOTE: During PDMS polymerization, curing agent crosslinks with the base elastomer to provide final polymer desired mechanical properties. Crosslink ratios can be adjusted to alter PDMS stiffness. - Place a drop (~0.2 mL) of desiccated PDMS mixture on a square or circular coverslip (e.g., 22 mm x 22 mm) and rotate the coverslip on a spin coater at 2500 rpm for 30 s.
- Incubate the coverslip in either an oven at 60 °C for 1-2 h or room temperature overnight for PDMS to solidify.
- Treat the coverslip using UV-Ozone cleaner for 7 min (UV wavelength: 185 nm and 254 nm) to increase the surface hydrophilicity. Place it into a sterilized 6-well plate.
- Before using for cell culture, incubate substrates in 70% ethanol for at least 30 min.
CAUTION: UV-Ozone cleaner can generate Ozone that is harmful to humans. Work in a chemical fume hood or with some form of ventilation. - Immerse coverslips in the protein solution (10 µg/mL collagen I, fibronectin, or laminin) for 60 min in a sterile incubator at 37 °C.
NOTE: Following UV-Ozone treatment, the PDMS surface may still be hydrophobic. Rotate the well plate to ensure each coverslip is covered with the protein solution. - Aspirate the protein solution and wash the coverslip with phosphate buffered saline (PBS) 3x.
- Re-suspend RT4-D6P2T Schwann cell line (SCs) from passaging dish using commercially available EDTA solution (1x) with 2.5% trypsin and count cells with hemocytometer. Seed SCs on the tunable PDMS surface at the desired cell density. SC seeding densities may vary for each different application.
- Maintain cells in the desired cell culture parameters (90% humidity, 5% CO2, 37 °C, etc.) for the length of experiment. Use Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin as the cell culture medium.
- Mix the PDMS base elastomer and curing agents using a pipette tip vigorously at a ratio between 10:1 and 60:1 until bubbles are homogeneously dispersed within the mixture. Remove bubbles using vacuum desiccation until bubbles are dissipated.
- Micropatterned substrate preparation
- Draw the desired geometry and cell adhesive areas (900 µm2, 1,600 µm2 and 2,500 µm2) using computer-aided design (CAD) software. Create a chrome photomask based on those patterns from a commercial supplier.
- In a clean room or dust free environment, use standard photolithography techniques to fabricate silicon wafers (protocols are detailed elsewhere31). Critical parameters for this particular application are as follows: Photoresist: SU-8 2010; Spin profile to disperse the photoresist: 500 rpm for 10 s with an acceleration of 100 rpm/s, then 3500 rpm for 30 s with an acceleration of 300 rpm/s; Exposure energy of UV light: 130 mJ/cm2.
NOTE: The height of patterns on the silicon wafers is approximately 10 µm following these parameters. Potential cracks around the edge outside of the rectangular or triangular patterns can be seen using a light microscope after step 1.2.2. Baking the silicon wafer at 190 °C for 30 min helps to eliminate the cracks. - Place the patterned silicon wafer inside a circular 150 mm diameter x 15 mm height Petri dish and pour de-gassed PDMS (mixing ratio 10:1) as prepared in step 1.1.1 onto the silicon wafer.
NOTE: Ensure the thickness of PDMS is at least 5 mm for ease of handling during microcontact printing steps. - Solidify PDMS on silicon wafer in an oven at 60 °C overnight. Allow PDMS to cool to room temperature. Precisely cut stamps of 30 mm x 30 mm squares containing the correct patterns from the silicon wafer using a surgical scalpel. Do not damage the silicon wafer.
NOTE: Silicon wafers can be reused many times at this point to produce more stamps following cleaning with isopropanol. - Sterilize PDMS stamps and the tunable coverslips (prepared in step 1.1.1 to 1.1.3) by immersing them into 70% ethanol for 30 min.
- To confirm the efficacy of micropattern by PDMS stamps after microcontact printing, dry the surface of PDMS stamps using a filtered air stream and pipette 50 µg/mL BSA (Texas Red conjugated) solution to cover the entire patterned side of the PDMS stamp.
- Incubate PDMS stamps with BSA solution for 1 h at room temperature to allow for protein adsorption.
- Dry the surface of tunable coverslips using a filtered air stream, increase surface hydrophilicity as described in step 1.1.5.
- Air dry the PDMS stamps to remove the remaining BSA solution.
NOTE: Take care that BSA solution is completely removed from the stamp because any remaining solution will cause stamps to slide on the coverslip during microcontact printing. - Bring the patterned side of the stamp into conformal contact with the tunable coverslip for BSA adsorption on the coverslip surface. Gently press the stamp against the coverslip for 5 min.
NOTE: Do not apply excessive force on the stamp since it will bend and cause non-specific contact between stamp and coverslip. The appropriate amount of force applied on the stamp is essential for successful microcontact printing. - Examine the micropattern using fluorescence microscope with a FITC (Fluorescein isothiocyanate) filter.
- To print cell adhesive areas rather than fluorescent patterns, substitute laminin for BSA protein and repeat step 1.2.5 to 1.2.10.
- Remove stamps from coverslips, transfer coverslips into a sterilized 6-well plate. Add 2 mL of 0.2% w/v Pluronic F-127 solution into each well to cover the surface of the coverslip and incubate for 1 h at room temperature.
NOTE: Pluronic F-127 can be adsorbed to PDMS surface increasing the hydrophobicity of PDMS surface to block cells from adhesion. - Aspirate Pluronic F-127 solution and wash 5x with PBS and 1x with the cell culture medium before seeding cells. A typical seeding density for SCs is 1,000 cells/cm2.
- 45 min following cell seeding, remove the cell culture medium and wash coverslips with PBS 2x to prevent multiple SCs from adhering to the same pattern. Maintain cells in desired cell culture environment for 48 h before quantification.
- To create line-patterned cell culture substrates to examine aligned cells, follow step 1.2.1 to 1.2.4 to create stamps for microcontact printing.
NOTE: The dimensions of the groove/ridge of the lined patterns on stamp is 50 μm x 50 μm for the cells outlined. The total dimensions of the stamp are 10 mm x 10 mm. - Cut stamps into dimensions containing only desired line patterns.
NOTE: When creating stamps in CAD, the unpatterned area of the stamp around the line patterns will correspond to cell adhesive area following microcontact printing. Thus, it is necessary to eliminate unpatterned areas when cutting out the stamp to ensure every SC seeded on the surface follows the patterns. - Follow step 1.1.1 to 1.1.3 to prepare tunable PDMS surface coating two Petri dishes.
NOTE: This will be PDMS covering the Petri dish surface itself and not on a coverslip. - Follow step 1.2.5 to 1.2.10 to perform microcontact printing to print line-patterned cell adhesive areas on one of the PDMS coated Petri dishes.
NOTE: The surface area of a 60 mm x 15 mm Petri dish can contain line-patterned areas from 6 PDMS stamps. - Remove stamps and fill the Petri dish with 4 mL of 0.2% w/v Pluronic F-127 solution and incubate for 1 h.
- After microcontact printing, rinse each side of the PDMS stamps with 70% ethanol 3x and dry with air. Rotate PDMS stamps and follow step 1.2.5 to 1.2.10 to print unpatterned cell adhesive area using the unpatterned side of the stamp on the second Petri dish. Repeat step 1.2.13.
- Aspirate F-127 solution from dishes, wash 3x with PBS followed with 1x wash using fresh cell culture medium. Seed SCs on dishes.
NOTE: The seeding density for a line-patterned dish is 5,000 cells/cm2 and for an unpatterned dish is 10,000 cells/cm2. - Maintain SCs at desired conditions for 48 h and follow protocols to prepare SC lysates32.
NOTE: The cell seeding density for unpatterned dishes is 2x higher than that for line-patterned dishes, as the line-patterned dish has only half the cell adhesive area of the unpatterned dish.- To prepare lysates, transfer adequate radioimmunoprecipitation assay (RIPA) buffer to a 10 mL conical centrifuge tube. Dilute protease and phosphatase inhibitor (100x) at a ratio of 1:100 in RIPA buffer, mix well by pipetting.
- Wash cells with ice cold PBS (1x) for 2 min, add 80 µL of the solution prepared from step 1.2.23.1 onto each cell adhesive area (the area that contacted with PDMS stamps and adsorbed protein) within petri dishes. Incubate cells with the solution on an ice block for 15 min.
NOTE: The solution will only stay on cell adhesive area due to the hydrophobicity of Pluronic F-127 adsorption elsewhere. This feature enables a successful and sufficient protein extraction for line patterned SCs. - Scrape SCs with a cell scraper for 5 min. Collect lysate into a labeled 1.5 mL microcentrifuge tube.
- Microcentrifuge lysate at 12, 000 x g for 15 min at 4°C. Collect supernatant with a 1,000 µL pipette and transfer to a clean microcentrifuge tube. Store cell lysate at -20 °C.
- Substrate characterization
NOTE: To characterize mechanics of the polymer on the coverslip, multiple methods are generally employed including bulk compression testing11,33 or atomic force microscopy testing34. This protocol will outline bulk compression testing.- Pour the PDMS precursor of the desired mixing ratio (step 1.1) into a 30 mm Petri dish, ensure the thickness of the PDMS layer within the Petri dish is at least 20 mm.
- Remove the Petri dish with solidified PDMS from 60 °C oven after 1 h and allow to cool at room temperature. Cut polymer into 10 mm x 10 mm squares. Measure the thickness of PDMS using calipers.
- Place PDMS on stage of the compression force measuring machine. Plug in compression force sensor (model: 112C) to sensor port and fix the sensor to the axis of the test machine.
- Adjust the height of the sensor to approximately 0.5 cm above the PDMS stamp using “jog” control on front panel of instrument.
- Open the associated software using “Test Setup” window, select “Servo profile”, and open the “Segment” window. In the “Segment” window, input desired “Control Rate” and “End Amount” for the test.
NOTE: Control rate determines the rate at which the sensor travels down towards the PDMS. End amount determines the total distance the sensor travels. - Use the “Z” button located on control panel of the software to reset all measurements at this point.
- Move the sensor down to lightly contact PDMS until 1-2 newtons (N) are loaded. The loading and the distance that the sensor travels will be displayed in the software.
- After utilizing the “Z” button, run measurement using “Play”, and save the file recording force and distance.
- Repeat steps 1.3.3 to 1.3.8 for each experimental condition of PDMS.
- Open the file and use the following formula to calculate the Young’s modulus (E) of PDMS for each ratio. (F = compression force, A = area of PDMS stamp, ∆L = traveling distance of sensor, and L0 = original thickness of the PDMS stamp).
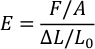
2. Quantification of cellular properties on tunable substrates
- Proliferation assay
- Seed SCs on substrates prepared from step 1.1.9 at a density of 5,000 cells/cm2 in a 6-well plate. Allow SCs to incubate for 48 h in standard cell culture conditions (37 °C and 5% CO2).
- Dilute 12 µL of 10 mM Bromodeoxyuridine (BrdU) stock solution into 12 mL of 37 °C cell culture medium, mix well with pipette to make 10 µM BrdU labeling solution.
- Remove the cell culture medium and wash SCs 2x with PBS.
- Add 2 mL of BrdU labeling solution into each well and incubate SCs for 2 h.
NOTE: The incubation time of BrdU labeling solution depends on the specific cell proliferation rate. The RT4-D6P2T SC line has a high proliferation rate so 2 h of incubation time was used. - Remove BrdU labeling solution and wash SCs 3x with PBS. Add 1 mL of 3.7% formaldehyde in PBS to each well and incubate at room temperature for 15 min for cell fixation.
CAUTION: Formaldehyde is a human carcinogen; therefore, carry out all work inside a chemical fume hood with appropriate protection.
NOTE: When washing with PBS, there is no cell culture medium in the well, and thus the PDMS surface may be hydrophobic. Take precautions to not completely dry the surface of the substrate to prevent cell damage. - Aspirate formaldehyde solution and wash 3x with PBS (3 min each). Remove PBS and add 1 mL of 0.2% Triton X-100 in PBS to each well to permeabilize cell membrane. Incubate SCs with Triton X-100 solution for 20 min at room temperature.
- Remove Triton X-100 solution and wash SCs 3x with PBS (3 min each).
- Add 1 mL of 1 N HCl into each well and incubate on ice for 10 min. Remove 1 N HCl and add 1 mL of 2 N HCl into each well and incubate at room temperature for 10 min. HCl treatment is for DNA hydrolysis.
- Mix 182 mL of 0.2 mM Na2HPO4 and 18 mL of 0.1 mM citric acid to produce a phosphate/citric acid buffer for antigen retrieval. Remove 2 N HCl and add 1 mL phosphate/citric acid buffer into each well and incubate at room temperature for 10 min.
- Wash SCs 3x with 0.2% Triton X-100 in PBS. Add 2 mL of 3% bovine serum albumin (BSA) in PBS into each well and incubate for 30 min at room temperature to clock nonspecific binding of the antibody.
- Dilute BrdU primary antibody conjugated with Alexa Fluor 488 in 3% BSA solution at a ratio of 1:300 for BrdU staining solution. Incubate SCs with staining solution overnight at room temperature while plate is covered in aluminum foil.
- To quantify proliferation, image SCs using the FITC and DAPI channel of a fluorescent microscope to detect BrdU and nuclei, respectively. Save images as “nd.2” files.
- Open “nd.2” files for each image taken at identical spatial positions.
- Open the image analysis software. Right click the background to open the window “Automated Measurement Results” and “Automated Measurement” in the section of “Analysis Control”.
- In “Count & Taxonomy” menu, select “Count”. On FITC image, click on each nucleus showing green fluorescence (BrdU positive) and right click on the image.
NOTE: The number of BrdU positive cells are shown in the window of “Automations and Measurements”. - For DAPI images, repeat step 2.1.14 to count the number of total nuclei. Calculate the percentage of BrdU positive cells for this image.
- Repeat step 2.1.13 through 2.1.16 for other images for statistical purposes and calculate the mean percentage of BrdU positive cells for each substrate condition.
- Quantification of c-Jun expression through immunofluorescent image analysis
- Cells prepared inside 6-well plates from step 1.1.9 and 1.2.23 are fixed and permeabilized with the procedures previously described (step 2.1.5-2.1.7).
NOTE: To perform accurate comparisons of fluorescent intensity across cells of differing ECM conditions, apply camera settings identically across all samples with all samples having the same parameters. - Save images as “.nd2” files.
- Open the image analysis software. Right click background to open the window “Automated Measurement Results” and “Automated Measurement” in the section of “Analysis Control”.
- In “Automated Measurement Results”, select “Object Data”. Activate “Keep updating measurement” button.
- In the top panel of the software, select “Measure” followed by “Object features”. Add “Mean Intensity” to the section of “Selected for Measurement”.
- Open and merge two “.nd2” image files that contain images of c-Jun and nuclei.
- In the top panel, select “ROI” and select “Draw Rectangular ROI”. Draw a rectangular area containing the nuclear area of a single cell.
NOTE: c-Jun expression is concentrated within nuclei35. - In top panel of software, select “Binary” and “Define Threshold”, a new window will appear to precisely define c-Jun fluorescent area.
- In new window, click “Full Image/Use ROI” to switch program from full image model to the ROI model. Use “Intensity” to adjust the lookup table located on the left side of the window for adjustment of size/shape of the highlighted area within the rectangular ROI.
NOTE: Take care to ensure the size/shape of the highlighted area is identical to the nucleus. - Click the “OK” button, to get the mean FITC intensity in the window of “Automated Measurement Results” and then click “Store Data”.
- In the window of “Automated Measurement”, “Delete Object” to remove red highlighted area. On the left side panel, use “Pointing Tool” to select the rectangular ROI and delete.
- Repeat step 2.2.6 to 2.2.11 to measure mean FITC intensity for each additional cell.
- In window area of “Automated Measurement Results”, select “Stored” and all stored data will be presented. Use the “Export” function and select “Data to Excel” to save the exported spreadsheet and perform additional calculations.
- Cells prepared inside 6-well plates from step 1.1.9 and 1.2.23 are fixed and permeabilized with the procedures previously described (step 2.1.5-2.1.7).
- Quantifying nuclear elongation
- Fix and permeabilize SCs prepared from step 1.2.22 following steps 2.1.5-2.1.7. Perform nuclear staining using mounting medium with DAPI.
- Using the DAPI channel and a 40x objective lens, acquire images of the sample and save as “.nd2” files.
- Follow step 2.2.3 and 2.2.4 to open “Automated Measurement Results” and “Automated Measurement” window in the image analysis software.
- In “Automated Measurement Results”, use the “Option” function followed by “Select Object Feature”. In the “Feature” column, select “Elongation” and add to “Selected for Measurements” column. Use “Keep Updating Measurement” to activate this function.
- Open the “nd.2” image file that contains the nuclear images. In “Automated Measurement” window, select “Auto Detect” function and select a nucleus. Right click on the image and the measured nuclear aspect ratio will be shown in “Automated Measurement Results”.
- Repeat step 2.3.5 to quantify nuclear aspect ratios for other nuclei within the image. Select “Store Data” in the window of “Automated Measurement Results”.
- Repeat 2.3.5 to 2.3.6 for additional images. Export the data to a spreadsheet file as previously done in 2.2.13 for analysis.
- Western blot to quantify protein expression
- Follow standard protocols for western blot analysis detailed elsewhere32. The dilutions of antibodies used in the study are shown below: Rabbit anti c-Jun 1:2,000; Mouse anti β-actin 1:1,000; Rabbit anti p75NTR 1:1,000; Rabbit anti myelin basic protein 1:1,000; Anti-mouse/rabbit IgG, HRP-linked antibody 1:10,000.
Access restricted. Please log in or start a trial to view this content.
Results
To analyze and quantify the interplay between substrate stiffness and protein composition on SC phenotype, a tunable PDMS cell culture substrate was developed (Figure 1A). Compression testing of the polymer at differing base: curing agent ratios was utilized to quantify the Young’s modulus (E) of the substrate (Figure 1B). The resulting range of modulus values represents physiologically relevant substrate conditions. Following preparation of substrates, SC...
Access restricted. Please log in or start a trial to view this content.
Discussion
SCs can promote nerve regeneration due to their phenotypic transformation and regenerative potential following nerve injury. However, how ECM cues regulate this regenerative capacity remains mostly unclear, potentially hindering not only the development of biomaterials that aim to promote nerve regeneration but also the understanding of the mechanisms involved in nerve regeneration. To begin to examine this interplay, cell culture substrates were created where ECM cues such as stiffness, protein coating, and adhesive top...
Access restricted. Please log in or start a trial to view this content.
Disclosures
No potential conflict of interest was reported by the authors.
Acknowledgements
The authors gratefully acknowledge funding support from the University of Cincinnati. The authors also thank Ron Flenniken of the University of Cincinnati Advanced Materials Characterization laboratory for support.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Albumin from Bovine Serum (BSA), Texas Red conjugate | Thermo Fisher Scientific | A23017 | BSA staining to show micropatterns |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | 7076S | Antibody used for western blot analysis |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | 7074S | Antibody used for western blot analysis |
| BrdU | Thermo Fisher Scientific | B23151 | Reagent used to measure cell proliferation |
| BrdU primary antibody conjugated with Alexa Fluor 488 | Thermo Fisher Scientific | B35130 | Used to visualize BrdU in cell proliferation assays |
| Collagen I | Thermo Fisher Scientific | A10483-01 | Protein used to coat coverslips |
| Compression force test machine | TestResources | Instrument to quantify mechanical properties of polymers | |
| Dulbecco's Modified Eagle Medium | Thermo Fisher Scientific | 11965092 | Cell culture medium |
| Fetal Bovine Serum | Thermo Fisher Scientific | 16000044 | Cell culture medium supplemental |
| Fibronectin | Thermo Fisher Scientific | 33010-018 | Protein used to coat coverslips |
| Fluorescence microscope | Nikon | Eclipse Ti2 | Fluorescence microscope |
| Halt Protease and Phosphatase Inhibitor Cocktail (100X) | Thermo Fisher Scientific | 78440 | Protease and Phosphatase Inhibitor |
| Laminin | Thermo Fisher Scientific | 23017015 | Protein used to coat coverslips |
| Mounting medium with DAPI | Thermo Fisher Scientific | P36971 | Coverslip mountant and nuclei staining |
| Mouse c-Jun primary antibody | Thermo Fisher Scientific | 711202 | Primary antibody to visualize c-Jun protein |
| Mouse β-Actin primary antibody | Cell Signaling Technology | 3700S | Loading control for western blot experiments |
| Penicillin-Streptomycin | Thermo Fisher Scientific | 15140122 | Cell culture medium supplemental |
| Photoresist SU 2010 | KAYAKU | SU8-2010 | Photoresist |
| Pluronic F-127 | Sigma Aldrich | P-2443 | Block non-specific protein binding |
| Rabbit c-Jun primary antibody | Cell Signaling Technology | 9165S | Primary antibody for visualization of c-Jun protein |
| Rabbit myelin basic protein primary antibody | Abcam | ab40390 | Primary antibody for visualization of MBP |
| Rabbit p75NTR primary antibody | Cell Signaling Technology | 8238S | Primary antibody for visualization of p75NTR |
| Rhodamine phalloidin | Thermo Fisher Scientific | R415 | Visualization of cell cytoskeleton |
| RIPA buffer | Abcam | ab156034 | Cell lysis buffer |
| RT4-D6P2T Schwann cell line | ATCC | CRL-2768 | Cell line used in experiments |
| SYLGARD 184 PDMS base and curing agent | Sigma Aldrich | 761036 | Tunable polymer used to coat coverslips |
| Trypsin | Thermo Fisher Scientific | 15090-046 | Cell dissociation reagent |
| UV-Ozone cleaner | Novascan | Increase hydrophicility of PDMS | |
| Versene (1x) | Thermo Fisher Scientific | 15040066 | Cell dissociation reagent |
References
- Taylor, C. A., Braza, D., Rice, J. B., Dillingham, T. The Incidence of Peripheral Nerve Injury in Extremity Trauma. American Journal of Physical Medicine & Rehabilitation. 87, 381-385 (2008).
- Noble, J., Munro, C. A., Prasad, V. S. S. V., Midha, R. Analysis of Upper and Lower Extremity Peripheral Nerve Injuries in a Population of Patients with Multiple Injuries. Journal of Trauma and Acute Care Surgery. 45, 116-122 (1998).
- Jessen, K. R., Mirsky, R. The repair Schwann cell and its function in regenerating nerves. Journal of Physiology. 594, 3521-3531 (2016).
- Arthur-Farraj, P. J., et al. c-Jun Reprograms Schwann Cells of Injured Nerves to Generate a Repair Cell Essential for Regeneration. Neuron. 75, 633-647 (2012).
- Jessen, K. R., Mirsky, R. The Success and Failure of the Schwann Cell Response to Nerve Injury. Frontiers in Cell Neurosciences. 13, 33(2019).
- Gomez-Sanchez, J. A., et al. After Nerve Injury, Lineage Tracing Shows That Myelin and Remak Schwann Cells Elongate Extensively and Branch to Form Repair Schwann Cells, Which Shorten Radically on Remyelination. Journal of Neuroscience. 37 (37), 9086-9099 (2017).
- Deumens, R., et al. Repairing injured peripheral nerves: Bridging the gap. Progress in Neurobiology. 92, 245-276 (2010).
- Höke, A., Gordon, T., Zochodne, D. W., Sulaiman, O. A. R. A decline in glial cell-line-derived neurotrophic factor expression is associated with impaired regeneration after long-term Schwann cell denervation. Experimental Neurology. 173, 77-85 (2002).
- Jones, S., Eisenberg, H. M., Jia, X. Advances and future applications of augmented peripheral nerve regeneration. International Journal of Molecular Sciences. 17, 1-17 (2016).
- Harris, G. M., et al. Nerve Guidance by a Decellularized Fibroblast Extracellular Matrix. Matrix Biology. 60-61, 176-189 (2017).
- Harris, G. M., Piroli, M. E., Jabbarzadeh, E. Deconstructing the Effects of Matrix Elasticity and Geometry in Mesenchymal Stem Cell Lineage Commitment. Advanced Function Mater. 24 (16), 2396-2403 (2014).
- Pryzhkova, M. V., Harris, G. M., Ma, S., Jabbarzadeh, E. Patterning pluripotent stem cells at a single cell level. Journal of Biomaterials and Tissue Engineering. 3 (4), 461-471 (2013).
- Engler, A. J., Sweeney, H. L., Discher, D. E., Schwarzbauer, J. E. Extracellular matrix elasticity directs stem cell differentiation. Journal of Musculoskeleton and Neuronal Interaction. 7 (4), 335(2007).
- Chen, C. S., Mrksich, M., Huang, S., Whitesides, G. M., Ingber, D. E. Geometric control of cell life and death. Science. 276 (5317), 1425-1428 (1997).
- Engler, A. J., Sen, S., Sweeney, H. L., Discher, D. E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 126, 677-689 (2006).
- Pickup, M. W., Mouw, J. K., Weaver, V. M. The extracellular matrix modulates the hallmarks of cancer. EMBO Reports. 15, 1243-1253 (2014).
- Chernousov, M. A., Carey, D. J. Schwann cell extracellular matrix molecules and their receptors. Histology and Histopathology. 15, 593-601 (2000).
- Shibata, S., et al. Selective Laminin-Directed Differentiation of Human Induced Pluripotent Stem Cells into Distinct Ocular Lineages. Cell Reports. 25 (6), 1668-1679 (2018).
- Mcbeath, R., Pirone, D. M., Nelson, C. M., Bhadriraju, K., Chen, C. S. Cell Shape, Cytoskeletal tenstion and RhoA regulate stem cell lineage committment. Developmental Cell. 6, 483-495 (2004).
- Halder, G., Dupont, S., Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nature Reviews Molecular Cell Biology. 13, 591-600 (2012).
- Jessen, K. R., Mirsky, R. The repair Schwann cell and its function in regenerating nerves. Journal of Physiology. 594 (13), 3521-3531 (2016).
- Lopez-Fagundo, C., Bar-Kochba, E., Livi, L. L., Hoffman-Kim, D., Franck, C. Three-dimensional traction forces of Schwann cells on compliant substrates. Journal of The Royal Society Interface. 11, 20140247(2014).
- Gu, Y., et al. The influence of substrate stiffness on the behavior and functions of Schwann cells in culture. Biomaterials. 33, 6672-6681 (2012).
- Xu, Z. Y., Orkwis, J. A., DeVine, B. M., Harris, G. M. Extracellular matrix cues modulate Schwann cell morphology, proliferation, and protein expression. Journal of Tissue Engineering and Regenerative. , (2019).
- Urbanski, M. M., et al. Myelinating glia differentiation is regulated by extracellular matrix elasticity. Scientific Reports. 6, 1-12 (2016).
- Sun, Y., et al. Tunable stiffness of graphene oxide/polyacrylamide composite scaffolds regulates cytoskeleton assembly. Chemical Sciences. 9 (31), 6516-6522 (2018).
- Hwang, J. H., et al. Extracellular matrix stiffness regulates osteogenic differentiation through MAPK activation. PLoS One. 10, 1-16 (2015).
- Ryan, A. J., et al. A Physicochemically Optimized and Neuroconductive Biphasic Nerve Guidance Conduit for Peripheral Nerve Repair. Advanced Healthcare Materials. 6, 1-13 (2017).
- Du, J., et al. Prompt peripheral nerve regeneration induced by a hierarchically aligned fibrin nanofiber hydrogel. Acta Biomaterialia. 55, 296-309 (2017).
- Huang, L., et al. A compound scaffold with uniform longitudinally oriented guidance cues and a porous sheath promotes peripheral nerve regeneration in vivo. Acta Biomaterialia. 68, 223-236 (2018).
- Brower, K., White, A. K., Fordyce, P. M. Multi-step Variable Height Photolithography for Valved Multilayer Microfluidic Devices. Jouranl of Visualized Experiments. (119), e55276(2017).
- Gupta, R., et al. Shear stress alters the expression of myelin-associated glycoprotein (MAG) and myelin basic protein (MBP) in Schwann cells. Journal of Orthopaedic Research : Official Publication of the Orthopaedic Research Society. 23, 1232-1239 (2005).
- Harris, G. M., Shazly, T., Jabbarzadeh, E. Deciphering the combinatorial roles of geometric, mechanical, and adhesion cues in regulation of cell spreading. PLoS One. 8 (11), (2013).
- Engler, A. J., Sen, S., Sweeney, H. L., Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell. 126 (4), 677-689 (2006).
- Schreck, I., et al. C-Jun localizes to the nucleus independent of its phosphorylation by and interaction with JNK and vice versa promotes nuclear accumulation of JNK. Biochemical and Biophysical Research Communications. 407, 735-740 (2011).
- Shen, K., Qi, J., Kam, L. C. Microcontact printing of proteins for cell biology. Journal Visualized Experiments. (22), e1065(2008).
- Treter, J., et al. Washing-resistant surfactant coated surface is able to inhibit pathogenic bacteria adhesion. Applied Surface Science. 303, 147-154 (2014).
- Lutz, J. F. Polymerization of oligo(ethylene glycol) (meth)acrylates: Toward new generations of smart biocompatible materials. Journal of Polymer Science Part A: Polymer Chemistry. 46 (11), 3459-3470 (2008).
- Marcus, M., et al. Interactions of Neurons with Physical Environments. Advanced Healthcare Materials. 6, (2017).
- Pu, J. Golgi polarization in a strong electric field. Journal of Cell Science. 118, 1117-1128 (2005).
- Blaker, J. J., et al. Bioactive Silk-Based Nerve Guidance Conduits for Augmenting Peripheral Nerve Repair. Advanced Healthcare Materials. 7, 1800308(2018).
- Daly, W., Yao, L., Zeugolis, D., Windebank, A., Pandit, A. A biomaterials approach to peripheral nerve regeneration : bridging the peripheral nerve gap and enhancing functional recovery. Journal of the Royal Society of Interface. 9 (67), 202-221 (2012).
- Xia, H., et al. Directed neurite growth of rat dorsal root ganglion neurons and increased colocalization with Schwann cells on aligned poly(methyl methacrylate) electrospun nanofibers. Brain Research. 1565, 18-27 (2014).
- Wang, H. B., Mullins, M. E., Cregg, J. M., McCarthy, C. W., Gilbert, R. J. Varying the diameter of aligned electrospun fibers alters neurite outgrowth and Schwann cell migration. Acta Biomaterialia. 6, 2970-2978 (2010).
- Carvalho, C. R., Oliveira, J. M., Reis, R. L. Modern Trends for Peripheral Nerve Repair and Regeneration: Beyond the Hollow Nerve Guidance Conduit. Frontiers in Bioengineering and Biotechnology. 7, 337(2019).
- Yang, Y., Wang, K., Gu, X., Leong, K. W. Biophysical Regulation of Cell Behavior - Cross Talk between Substrate Stiffness and Nanotopography. Engineering. 3, 36-54 (2017).
- Tan, J. L., Liu, W., Nelson, C. M., Raghavan, S., Chen, C. S. Simple Approach to Micropattern Cells on Common Culture Substrates by Tuning Substrate Wettability. Tissue Engineering. 10, 865-872 (2004).
- Grove, M., et al. YAP/TAZ initiate and maintain schwann cell myelination. Elife. 6, 1-27 (2017).
- Poitelon, Y., et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nature Neuroscience. 19, 879-887 (2016).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved