Method Article
Sample Preparation for Metabolic Profiling using MALDI Mass Spectrometry Imaging
* These authors contributed equally
In This Article
Summary
The goal of this protocol is to provide detailed guidance on the sample preparation when planning for experiments using MALDI MSI to maximize metabolic and molecular detection in biological samples.
Abstract
Metabolomics, the study to identify and quantify small molecules and metabolites present in an experimental sample, has emerged as an important tool to investigate the biological activities during development and diseases. Metabolomics approaches are widely employed in the study of cancer, nutrition/diet, diabetes, and other physiological and pathological conditions involving metabolic processes. An advantageous tool that aids in metabolomic profiling advocated in this paper is matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI). Its ability to detect metabolites in situ without labeling, structural modifications, or other specialized reagents, such as those used in immunostaining, makes MALDI MSI a unique tool in advancing methodologies relevant in the field of metabolomics. An appropriate sample preparation process is critical to yield optimal results and will be the focus of this paper.
Introduction
Metabolites, the intermediates or end products of metabolism, including nucleotides, amino or organic acids, lipids, are key components to biological functions and processes. Metabolomics, the study of metabolites, allows for the exploration of their biochemical interactions and the understanding of their roles in the context of basic, translational, and clinical research. Metabolites are strongly associated with the phenotypes of organisms and provide information on biochemical activities that occur during cellular metabolism1. Therefore, in addition to genomics and proteomics, metabolomics has emerged as an important tool in understanding both physiological and pathological conditions. For instance, metabolomics is used in elucidating the mechanisms behind existing drugs as well as their tolerance. In drug development, xenobiotic metabolism is useful in assessing the activity or toxicity of metabolites across species, which later translates to supporting personalized medicine2. Despite the broad application of metabolomics, imaging of metabolites can be challenging due to metabolites' chemical reactivity, structural heterogeneity, and broad concentration range3. However, the concentrations of labile metabolites including high energy compounds, glucose, lactate, glycolytic, pentose shunt pathway, and TCA cycle intermediates, phospholipids, neurotransmitters, signaling compounds, can change within seconds and progress over minutes when tissue enzymes are active during tissue harvesting procedures, such as postmortem ischemia in brain harvesting4,5,6. To ensure accurate metabolomics data acquisition, appropriate and careful sample preparation is critical7,8. Current established platforms for measuring metabolites include NMR, enzyme assays, and mass spectrometry (including liquid and gas chromatography), the last of which is discussed further below.
MALDI-MSI is a state-of-the-art technique that allows for the analysis of complex samples through the detection of individual molecular species. MALDI MSI grants the benefit of being able to quickly and reproducibly measure various molecular compounds in biological samples. Mass spectrometry imaging further allows for the production of images that represent tissue biology based on its composite metabolites, and does so while preserving the spatial distribution of the metabolites in the sample9. MALDI's ability to detect analytes in a sample without the use of antibody labeling, structural modifications, or other specialized reagents, such as those used in immunostaining, coupled with its ability to monitor hundreds of molecules within a single experiment comprise just a few of the advantages MS imaging grants when it comes to metabolic profiling10,11. In addition to commonly used matrix such as 2,5-dihydroxybenzoic acid (DHB) and 9-aminoacridine (9-AA), recently discovered novel matrix N -(1-Naphthyl) Ethylenediamine Dihydrochloride (NEDC), which is well-suited for analyses of various low molecular weight metabolites, has further improved the application of MALDI MSI in metabolic profiling12.
Despite the broad application of MALDI MSI, the high cost of the instrument and the complexity of the experimental procedure prevents its wider implementation in individual research laboratories. Therefore, most of the MALDI MSI studies are supported through shared core facilities. The sample preparation, including slide preparation and matrix coating, is the most critical step in MALDI MSI. However, the slide preparation is normally carried out in individual researcher's laboratory, which creates potential variations in later MALDI MSI acquisition. Here we aim to provide a detailed protocol for the sample preparation of biological samples before proceeding to MALDI MSI measurements, and use a metabolomic profiling of developmental mouse brain as an example.
Protocol
The protocol follows the guidelines of City University of New York (CUNY) Advanced Science research Center (ASRC)'s institutional animal care and use committee (IACUC).
1. Harvest the tissue
- Prepare the aluminum boat. Prepare a rectangle aluminum 10cm x 20 cm, fold in the middle to make 10 cm double layer square. Label the sample information on one side, and fold the other side to form a boat with bottom surface about 4 cm x 4 cm.
- Precool the boat on the liquid nitrogen (LN2) in a Styrofoam box.
NOTE: If the boat is too small, the sample might fall out of the boat and become over frozen by directly contacting LN2, which may result in fragmentation during cryosectioning. - Euthanize the animal by cervical dislocation following institutional IACUC guidelines, immediately dissect out the tissue of interest.
- Keep the interval between animal anesthesia and snap freezing as short as possible, to minimize the alteration of metabolites during tissue harvesting, especially the labial metabolites in the brain due to postmortem ischemia.
NOTE: Perfusion the animal with phosphate-buffered saline (PBS) will increase the duration of ischemia, further altering the concentrations of labile metabolites and exaggerating the artifactual results. Therefore, perfusion or washing the tissue with PBS is not recommended, unless the contamination of blood or body fluid is of more concerned than metabolites deterioration or washout for individual project.
- Keep the interval between animal anesthesia and snap freezing as short as possible, to minimize the alteration of metabolites during tissue harvesting, especially the labial metabolites in the brain due to postmortem ischemia.
- Immediately place the tissue into the aluminum boat floating on liquid nitrogen, close the lid of Styrofoam box, and freeze for 2-10 min depending on the size of the tissue: 2 min, 5 min, 7 min for postnatal day 1 (P1), P21, P60 mouse brain, respectively, and 10 min for P60 rat brain.
NOTE: Do not freeze the tissue for prolonged time (e.g., 5 min for P1 mouse brain) as the over-frozen might lead to tissue fragmentation during cryosectioning. - Remove the boat by forceps, fold the foil to wrap the tissue, transport on dry ice and store at -80 °C for later use. If followed by immediate sectioning, carry the samples on dry ice to the cryostat.
NOTE: To better preserve the metabolites, it is preferred to store the sample in intact tissue and section it right before proceeding to MALDI imaging. The tissue can be stored in -80 °C for up to 24 months.
2. Cryosection the tissue
NOTE: Wear gloves at all times when handling the Indium tinoxide (ITO) slides. Avoid direct breathing onto the slide or wear masks (optional) to prevent the contamination of human saliva onto the tissue section.
- Before sectioning the tissue, gather the desired number of MALDI compatible ITO coated glass slides.
- Test the conductivity of the slide using a voltmeter set to resistance. Label the side where a resistance measurement is read: this will be the side that the tissue sections adhere to. Always place the slide on a clean paper towel to avoid contamination.
- Pre-cool the slides in a cryostat set to -20 °C.
- If tissue samples were removed from the -80 °C, let the tissue equilibrate in the cryostat chamber for about 45-60 min depending on the size of the tissue. If samples are removing from dry ice, equilibrate it for about 30 min.
- Thoroughly clean the cryostat with 70% ethanol. Pre-cool all the necessary tools including thin-tipped artist brush and forceps in the cryostat chamber.
- Set the temperature of the cryostat chamber and specimen head according to the type of the tissue: -14 °C for liver, -20 °C for muscle and -25 °C for skin9.
- Mount the tissue to the chuck using cryo tissue embedding compound OCT, avoiding OCT from the region of interest.
- Place a clean blade in the stage and lock. Adjust the position of the stage and the angle of the specimen to achieve the desired cutting angle.
NOTE: If different types/genotypes of tissue are to be cut, be sure to either reposition the sample so that a clean part of the blade is used , or change to a new blade before cutting the next sample to prevent cross-contamination. - Continue cutting until the region of interest (e.g., corpus callosum in the brain) is found. Be sure to keep the stage clean by brushing off extra pieces with an artist brush that has been equilibrated in the cryostat.
- Once the desired region is revealed, cut smaller sections of 10-12 µm thickness. If the section tends to flake or fall apart easily, raise the temperature of the cryostat, staying in the -22 °C to -11 °C range. We have found that the optimal cutting temperature for brain tissue is -15 °C to - 18 °C.
- Once a good section has been cut, adhere it to the ITO slide (operate in the cryostat chamber).
- Transfer one section of the tissue onto ITO slides using the tip of artist brush.
- Finger-warm the section by placing a finger under the slide to warm up the section to ensure secure mounting. The tissue section will first turn to transparent in 5-10 s and then turn opaque in about 30-60 s.
- Carefully set the slide aside in the cryostat.
- Repeat steps for other tissue samples, ensuring that each section of the tissue is placed evenly on the slide and is as aligned as possible.
- Since the MALDI target can hold up to two slides, place the sections from multiple samples of the same cohort onto a single slide or onto two slides, to ensure they can be analyzed at the same time.
- When finished, place ITO slides in a vacuum box and transfer to a desiccator with desiccant. Vacuum-dry the slide for 45-60 min.
NOTE: If a vacuum desiccator is not available in the lab, keep the slides under -20 °C all the time to avoid metabolites deterioration. - Slide storage and shipping: unless the sample is prepared by the MALDI imaging core facilities to proceed directly to MALDI imaging, store the slides at -80 °C or ship to core facilities or other MALDI research laboratories on dry ice.
- To best preserve the samples, place the slides into the slide transporter, fill it with nitrogen (optional), seal with parafilm, place into a zip bag, which is then placed into another zip bag containing desiccant. Label the outer zip bag.
- Proceed to storage at -80 °C (up to 6 months) or shipping with adequate dry ice.
3. Matrix Preparation
- Prepare the matrix.
NOTE: All the reagents must be HPLC grade.- Prepare NEDC at a concentration of 10 mg/mL. Prepare 10 mL of matrix dissolved in a solvent of 70% methanol (100 mg of NEDC, 7 mL of methanol, 3 mL of H2O).
- Additionally prepare an extra 10 mL of 70% methanol:water solution to flush the sprayer system before filling in the matrix.
- Once the slides are dehydrated in step 2.17, place "X" marks on the blank space of the glass slide outside of the tissue sections using a bold point silver marker, and then place a second "x" with a fine point black marker on top of the silver "X". The black "X" with a sharp contrast out of the silver background (the bold silver "X") will later serve as fiduciary marker for the later mass spectra acquisition in MALDI instrument.
- Load the slide into the MALDI slide metal target. Use plastic cover and draw/outline where the samples are onto the plastic cover. Set aside.
- Scan the image of the slide together with the MALDI target using flatbed scanner. The screws on the surface of the target will serve as the spacer to prevent the damage or contamination of the tissue section by the scanner. Preview and scan the selected slide area in 16-bit grayscale and 2400 dpi. Save the image for later use in MALDI MSI.
4. Matrix deposition
NOTE: There are multiple methods to apply an even layer of matrix in fine crystal size on to the MALDI slide, including sublimation, droplet inkjet printing, automatic matrix sprayer and manual spray using artist airbush9. We will use automatic matrix sprayer as an example in this protocol for its high reproducibility.
- Start up: Turn on the matrix sprayer unit, being sure that the valve is positioned at LOAD and launch the sprayer software. Check that the exhaust fan is operating well. Do not start the solvent pump if the active venting is not functioning properly.
- Confirm on the Comms tab that everything is communicating, and then start the solvent pump at .1 mL/min, with a backpressure of ~500 psi (or 3.4 MPa).
- Start compressed air flow to the matrix sprayer by setting the nitrogen tank to 30 psi. Then, adjust the pressure regulator on the front of the sprayer to 10 psi, and set sprayer nozzle temperature as desired.
NOTE: If the air pressure in prayer is lower than 5 psi, the sprayer nozzle will not be able to heat up for protection. - With the valve still in the LOAD position, use a syringe to flush the loop with 7 mL of 70% methanol, and then fill the loop with 6 mL of matrix.
- Place the tissue slides into the holders in the sprayer, taping down both ends to prevent movement, and to preserve a matrix-free edge to avoid the contamination of the metal clamp on MALDI target by the matrix.
NOTE: Testing matrix spray on a blank microscope slide first before proceeding to precious sample slides is highly recommended. - Check that the flow rate and temperature are stable to begin spraying.
- Select desired method pre-tested using blank glass slide.
- Press Start. This will set the nozzle temperature and adjust the pump flow rate to match the selected method. Switch the valve from Load to Spray position then confirm by clicking Continue.
- Allow the system to run, which will take 5-20 min per slide depending on the method. Switch the valve from Spray to Load position then confirm by clicking Continue when finished.
- Remove the slide(s) from the sprayer, examine the pattern of matrix coating under microscope to ensure an even layer of fine matrix crystal.
- After matrix deposition, place the slide(s) into the metal MALDI holder for immediate use. If there is only one specimen slide, add another blank slide to fill the two spaces of the MALDI holder.
- Clean up the system immediately after the usage following the manufacturer's instruction, to prevent the clogging of sprayer nozzle.
- After the sample slide is coated with matrix, either proceed immediately to MALDI imaging instrument, or ship it with dry ice using the same double zip bag preparation described in step 2.19.
NOTE: Under emergency circumstance such as the MALDI instrument is not available for immediate usage, store the coated slide in vacuum condition or at -20 °C for up to 24 hrs, although the deterioration of some metabolites might happen during the storage, which has not been thoroughly studied.
Results
The representative experiment was performed according to the workflow shown in Figure 1. The developmental C57BL wildtype mouse brains of postnatal day 1, 21, 60 (adult) were harvested as described above following CUNY IACUC guidelines and were snap frozen for 2, 5 and 7 min, respectively, on an aluminum boat floating on liquid nitrogen. The frozen tissues were cryosectioned at 10 µm thickness sections at −15 °C set for both specimen head and the chamber. The tissue cryosections were then gently transferred onto the pre-cooled conductive side of ITO-coated glass slides for MALDI imaging. Mounted cryosections on ITO slides were desiccated in vacuum for 45 min at room temperature, followed by matrix deposition using automatic matrix sprayer. Matrix NEDC was used to detect metabolites, and a matrix solution of 10 mg/mL in methanol/water (70/30, v/v) was deposited at a flow rate of 0.1 mL/min and a nozzle temperature of 75 °C for 12 cycles with 5 s drying between each cycle. A spray velocity of 1300 mm/min, track spacing of 2 mm, N2 gas pressure of 10 psi and flow rate of 3 L/min and nozzle height of 40 mm was used.
MALDI mass spectra were acquired in negative ion mode by MALDI time of flight (TOF) MSI instrument. 0.5-1 µL of red phosphorus (Pn clusters with n = 1 - 90) emulsion in methanol was deposited onto the ITO slides, next to the mounted tissues, and used to calibrate the instrument in the 100 - 1000 m/z mass range by applying quadratic calibration curve13. The laser spot diameters were focused to "Medium" modulated beam profile for 50 µm raster width. Spectra within the mass range from m/z 50 to 1000 were acquired at a 1000 Hz for 500 shots. Mass spectra data were recorded, and the imaging was further analyzed using advanced MALDI MSI data analysis software. Ion images were generated with root-mean square (RMS) normalization at a bin width of ±0.25 Da.
The results in Figure 2 show output images from MALDI MSI data analysis software of m/z spectra selected from every 100 Dalton interval, clearly depicting the utility for identification of spectra from small molecule metabolites to high molecular weight lipids. Each row depicts respective ion heat maps containing both spatial and spectral information of a certain metabolite species across three tissues collected at postnatal day 1, 21, and 60. For each representative m/z, analysis of regional distribution and ion abundances can be used to compare relative amounts of corresponding species between different ages. A strength of the MALDI MSI methodology is the ability to discern the specificity of certain species identified by m/z to developmental milestones or specific anatomical structures. Some metabolites are observed to be enriched in P1 neonates (m/z 320.1), enriched in P60 adults (m/z 846.5), or uniformly distributed across ages (m/z 480.3); other molecular species are specifically enriched in gray matter (m/z 117.1; 524.3; 765.1), white matter (m/z 673.4; 846.5), or CSF/ventricles (m/z 239.0) (Figure 2A). The spatial distribution of representative metabolites including hypoxanthine (m/z 135.0), N-Acetyl-L-aspartic acid (m/z 174.0), Arachidonic acid (m/z 303.2), and several lipids such as lysophosphatidylethanolamine LPE (18:1) (m/z 478.3), LPE(20:4) (m/z 500.3), LPE (22:6) (m/z 524.3), phosphatidylethanolamine PE (44:10) (m/z 838.5), Phosphatidylinositol PI(38:4) (m/z 885.6) are also shown (Figure 2B).
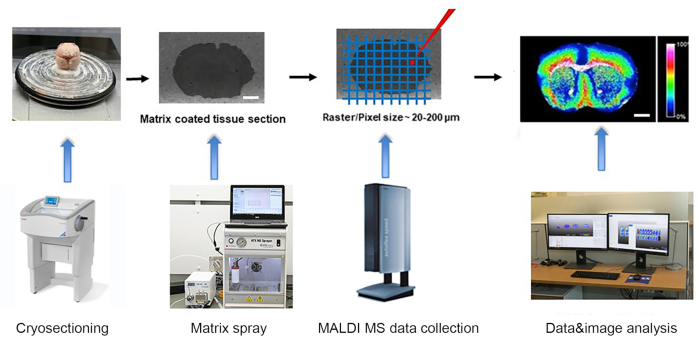
Figure 1. Workflow of MALDI- time of flight (TOF) mass spectrometry imaging. The snap frozen tissue is cryrosectioned in cryostat and mounted on ITO slide. → The slide with tissue sections is coated with a fine layer of matrix using automatic matrix sprayer. → Mass spectra is collected by MALDI-TOF MSI instrument at a raster of 20-200um. → The data is analyzed, and the images are generated using advanced MALDI MSI data analysis software. Scale bar: 2 mm. Please click here to view a larger version of this figure.
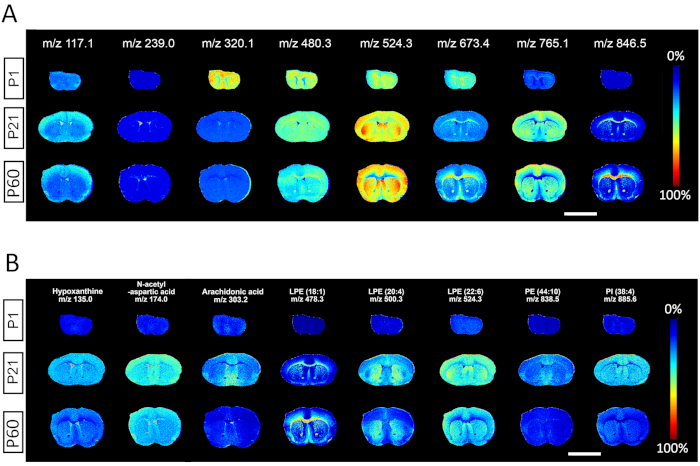
Figure 2. Representative output with selected m/z spectra from mass spectrometry acquired at 50 µm lateral resolution. (A) A heat map depicts the spatial distribution of specific metabolite species selected from every 100 Dalton m/z interval across the three developmental milestones identified at P1, P21 and P60. (B) The spatial distribution of representative metabolites at P1, P21 and P60, including: hypoxanthine, N-Acetyl-L-aspartic acid, Arachidonic acid, and different lipid species such as lysophosphatidylethanolamine (LPE), phosphatidylethanolamine (PE), Phosphatidylinositol (PI). Scale bar, 500 µm. Please click here to view a larger version of this figure.
Discussion
MALDI-Imaging (MALDI-MSI) is a label-free imaging technique that allows researchers to investigate the distribution of various biomolecules and their modifications in tissue, the molecular basis of pathology. Combined use of MALDI-MSI with traditional LC-MS approaches for tissue analysis provides the same molecular depth as traditional Omics workflows but which also retains the spatial relationship of those signals within the cellular network. The sample preparation is the most critical step in MALDI MSI and accounts for the variation in the final read outs of metabolomics studies carried out in different labs4. Here we provide a comprehensive yet practical protocol to standardize the sample preparation for metabolomic profiling using MALDI MSI, in the hope that it benefits a broad research community to implement MALDI MSI in their current and future research from basic biology to translational studies.
One must always bear in mind all precautions to minimize changes in the molecular profiles (both abundances and spatial distribution) during sample preparations and avoid contamination. Firstly, minimize the time between animal euthanasia and tissue harvesting, such as frozen in situ or heated by microwave fixation to inactivate enzymes in the brain to reduce the liability to postmortem ischemia4,5,6. Secondly, the snap freezing condition of the sample is critical. Inadequate freezing will cause the degradation and loss of the metabolites, while over freezing will lead to tissue fragmentation during cryosectioning. The freezing time should always be tested first according to previous reported studies, and the study of developmental mouse brain presented in this paper provides the reference points for rodent brain tissue. Thirdly, cutting of biological tissues and transferring their sections to the ITO slides will require adequate practice. It is important to note that while using the brush to pick up the section from the stage, the brush should be used delicately. Allow the bristles of brush to only come in contact with the edges of the tissue section to decrease the risk of contamination and section fragmentation. Flatten the section on the slide as much as possible, this will prevent the curling of the section during finger-warming. Fourthly, while mounting onto the ITO slide, make sure the entire section is well attached to the ITO slide as different regions of the tissue might require different time of finger-warming. For example, brain tumor tissue requires longer warming time than normal brain tissue. A poor mounting might lead to detachment and fragmentation of the tissue during MALDI-MS scanning. Bear in mind that the finger warming might enable enzyme action and metabolism causing artifactual changes of metabolites. Fifthly, a fine and uniform deposition of MALDI matrix plays an important role in achieving accurate spatial information and strong MALDI-MS signal. It is recommended to test the matrix spraying on a blank slide, and observe the crystal pattern under a microscope to verify proper coverage, before proceeding to the coating of precious specimen slide. And lastly, since an individual researcher executes the tissue harvesting and slide preparation at different speeds, it would be ideal to have one person to handle the sample preparation for the specimen in the same cohort, to minimize the variation.
The protocol provided above details standard procedures, which can be tailored towards the needs of particular experiments. For example, a cryo-sectioning gel OCT, which is normally used in cryo-sectioning of the sample, can be further utilized as a mounting glue to the tissue chuck (as in the study described above). Prior studies have shown that the polymer component in OCT causes strong ion suppression14. However, embedding the sample may be unavoidable in the cases where the tissue is too fragile to be cut without additional support from a polymer gel. In order to combat signal suppression in these cases, the tissues may need to be washed with serial washing in 70% ethanol and 95% ethanol to remove residual OCT for the detection of proteins or lipids, while the washing is not recommended for the detection of small molecular metabolites9.
MALDI MSI has grown increasingly relevant in both the research laboratory and clinical practice setting. For instance, MALDI MSI has recently proved useful in studies of proteomics in order to characterize the phenotypic functional status of an organism15, and in acting as an agent for microbial identification and diagnosis of subsequent disease16. While MALDI MSI supports a wide range of applications, there are some limitations associated with relying solely on this technique, especially when it comes to differentiating between similar species or metabolites, and the identification of specific targets. Another challenge is the quantification of the metabolites concentration according to MALDI MSI signals. It is often assumed that ion abundances in MALDI MSI spectra and the spatial distribution (or relative abundances) of corresponding molecular species across dissected tissues are well correlated. However, one should always bear in mind that the relationship between ion intensity and the amount of corresponding molecular species is complicated by numerous factors including, but not limited to, effects of ion suppression, changes in tissue structure and ion-molecule reactions17. Techniques that make use of internal standards can be implemented for absolute quantification (µmol/g tissue) in MALDI-MSI18. These two challenges are typically addressed with the combined workflow of MALDI MSI with liquid chromatography tandem MS(LC-MS/MS) techniques, whereby MALDI-MS allows for the mapping of the region of interest, which is later subjected to microextraction and LC-MS/MS to provide more information for identifying the metabolite19.
MS-based imaging methods have been developed in recent years as an alternative modality to previous techniques for imaging small molecule metabolites. With the advancements and growing popularity of MALDI MSI, it is expected that MALDI imaging will become a new standard tool for visualizing small molecules. Imaging of lipid and endogenous small molecules (e.g. neurotransmitters and metabolites) in the biological context, as well as imaging of xenobiotics for new pharmaceutical agents development are of particular interest. These three areas are expected to have rapid advancements with application of MALDI MSI in the near future20.
Disclosures
The authors declare no competing financial interests.
Acknowledgements
Ye He and Rinat Abzalimov are supported by the Professional Staff Congress-City University of New York (PSC-CUNY) Research Award Program. Yuki Chen and Kelly Veerasammy are supported by the Alfred P. Sloan Foundation CUNY Summer Undergraduate Research Program.
Materials
| Name | Company | Catalog Number | Comments |
| Andwin Scientific Tissue-Tek CRYO-OCT Compound | Fisher Scientific | 14-373-65 | |
| Artist brush MSC #5 1/8 X 9/16 TRIM RED SABLE | Fisher Scientific | 50-111-2302 | |
| Autoflex speed MALDI-TOF MS system | Bruker Daltonics Inc | MALDI-TOF MS instrument | |
| BD Syringe with Luer-Lok Tips | Fisher Scientific | 14-823-16E | |
| BD Vacutainer General Use Syringe Needles | Fisher Scientific | 23-021-020 | |
| Bruker Daltonics GLASS SLIDES MALDI IMAGNG | Fisher Scientific | NC0380464 | |
| Drierite, with indicator, 8 mesh, ACROS Organics | Fisher Scientific | AC219095000 | |
| Epson Perfection V600 Photo Scanner | Amazon | Perfection V600 | |
| Fisherbrand 5-Place Slide Mailer | Fisher Scientific | HS15986 | |
| Fisherbrand Digital Auto-Range Multimeter | Fisher Scientific | 01-241-1 | |
| FlexImaging v3.0 | Bruker Daltonics Inc | Bruker MS imaging analysis software | |
| HPLC Grade Methanol | Fisher Scientific | MMX04751 | |
| HPLC Grade Water | Fisher Scientific | W5-1 | |
| HTX M5 Sprayer | HTX Technologies, LLC | Automatic heated matrix sprayer | |
| Kimberly-Clark Professional Kimtech Science Kimwipes Delicate Task Wipers | Fisher Scientific | 06-666A | |
| MSC Ziploc Freezer Bag | Fisher Scientific | 50-111-3769 | |
| N -(1-Naphthyl) Ethylenediamine Dihydrochloride (NEDC) | Millipore Sigma Aldrich | 222488 | |
| SCiLS Lab (2015b) | SCiLS Lab | Advanced MALDI MSI data analysis software | |
| Thermo Scientific CryoStar NX50 Cryostat | Fisher Thermo Scientific | 95-713-0 | |
| Thermo Scientific Nalgene Transparent Polycarbonate Classic Design Desiccator | Fisher Scientific | 08-642-7 |
References
- Watkins, S. M., German, J. B. Metabolomics and biochemical profiling in drug discovery and development. Current Opinion in Molecular Therapeutics. 4, 224-228 (2002).
- Fernie, A. R., Trethewey, R. N., Krotzky, A. J., Willmitzer, L. Metabolite profiling: from diagnostics to systems biology. Nature Reviews Molecular Cell Biology. 5, 763-769 (2004).
- Theodoridis, G. A., Gika, H. G., Want, E. J., Wilson, I. D. Liquid chromatography-mass spectrometry based global metabolite profiling: a review. Analytica Chimica Acta. 711, 7-16 (2012).
- Dienel, G. A. Metabolomic and Imaging Mass Spectrometric Assays of Labile Brain Metabolites: Critical Importance of Brain Harvest Procedures. Neurochemistry Research. 45, 2586-2606 (2020).
- Dienel, G. A. Metabolomic Assays of Postmortem Brain Extracts: Pitfalls in Extrapolation of Concentrations of Glucose and Amino Acids to Metabolic Dysregulation In Vivo in Neurological Diseases. Neurochemistry Research. 44, 2239-2260 (2019).
- Wasek, B., Arning, E., Bottiglieri, T. The use of microwave irradiation for quantitative analysis of neurotransmitters in the mouse brain. Journal of Neuroscience Methods. 307, 188-193 (2018).
- Andres, D. A., et al. Improved workflow for mass spectrometry-based metabolomics analysis of the heart. Journal of Biological Chemistry. 295, 2676-2686 (2020).
- Lu, W., et al. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annual Review of Biochemistry. 86, 277-304 (2017).
- Norris, J. L., Caprioli, R. M. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chemical Reviews. 113, 2309-2342 (2013).
- Miura, D., et al. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Analytical Chemistry. 82, 9789-9796 (2010).
- Han, J., et al. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics. 4, 128-140 (2008).
- Wang, J., et al. MALDI-TOF MS imaging of metabolites with a N-(1-naphthyl) ethylenediamine dihydrochloride matrix and its application to colorectal cancer liver metastasis. Analytical Chemistry. 87, 422-430 (2015).
- Sladkova, K., Houska, J., Havel, J. Laser desorption ionization of red phosphorus clusters and their use for mass calibration in time-of-flight mass spectrometry. Rapid Communication in Mass Spectrometry. 19, 3114-3118 (2019).
- Schwartz, S. A., Reyzer, M. L., Caprioli, R. M. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. Journal of Mass Spectrometry. 38, 699-708 (2003).
- Greco, V., et al. Applications of MALDI-TOF mass spectrometry in clinical proteomics. Expert Review of Proteomics. 15, 683-696 (2018).
- Singhal, N., Kumar, M., Kanaujia, P. K., Virdi, J. S. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Frontiers in Microbiology. 6, 791 (2015).
- Hankin, J. A., Murphy, R. C. Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Analytical Chemistry. 82 (20), 8476-8484 (2010).
- Prentice, B. M., Chumbley, C. W., Caprioli, R. M. Absolute Quantification of Rifampicin by MALDI Imaging Mass Spectrometry Using Multiple TOF/TOF Events in a Single Laser Shot. Journal of the American Society for Mass Spectrometry. 28 (1), 136-144 (2017).
- Quanico, J., Franck, J., Wisztorski, M., Salzet, M., Fournier, I. Combined MALDI Mass Spectrometry Imaging and Parafilm-Assisted Microdissection-Based LC-MS/MS Workflows in the Study of the Brain. Methods in Molecular Biology. 1598, 269-283 (2017).
- Trim, P. J., Snel, M. F. Small molecule MALDI MS imaging: Current technologies and future challenges. Methods. 104, 127-141 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved