A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Size Exclusion Chromatography to Analyze Bacterial Outer Membrane Vesicle Heterogeneity
In This Article
Summary
Bacterial vesicles play important roles in pathogenesis and have promising biotechnological applications. The heterogeneity of vesicles complicates analysis and use; therefore, a simple, reproducible method to separate varying sizes of vesicles is necessary. Here, we demonstrate the use of size exclusion chromatography to separate heterogeneous vesicles produced by Aggregatibacter actinomycetemcomitans.
Abstract
The cell wall of Gram-negative bacteria consists of an inner (cytoplasmic) and outer membrane (OM), separated by a thin peptidoglycan layer. Throughout growth, the outer membrane can bleb to form spherical outer membrane vesicles (OMVs). These OMVs are involved in numerous cellular functions including cargo delivery to host cells and communication with bacterial cells. Recently, the therapeutic potential of OMVs has begun to be explored, including their use as vaccines and drug delivery vehicles. Although OMVs are derived from the OM, it has long been appreciated that the lipid and protein cargo of the OMV differs, often significantly, from that of the OM. More recently, evidence that bacteria can release multiple types of OMVs has been discovered, and evidence exists that size can impact the mechanism of their uptake by host cells. However, studies in this area are limited by difficulties in efficiently separating the heterogeneously sized OMVs. Density gradient centrifugation (DGC) has traditionally been used for this purpose; however, this technique is time-consuming and difficult to scale-up. Size exclusion chromatography (SEC), on the other hand, is less cumbersome and lends itself to the necessary future scale-up for therapeutic use of OMVs. Here, we describe a SEC approach that enables reproducible separation of heterogeneously sized vesicles, using as a test case, OMVs produced by Aggregatibacter actinomycetemcomitans, which range in diameter from less than 150 nm to greater than 350 nm. We demonstrate separation of "large" (350 nm) OMVs and "small" (<150 nm) OMVs, verified by dynamic light scattering (DLS). We recommend SEC-based techniques over DGC-based techniques for separation of heterogeneously sized vesicles due to its ease of use, reproducibility (including user-to-user), and possibility for scale-up.
Introduction
Gram-negative bacteria release vesicles derived from their outer membrane, so-called outer membrane vesicles (OMVs), throughout growth. These OMVs play important roles in cell-to-cell communication, both between bacteria and host as well as between bacterial cells, by carrying a number of important biomolecules, including DNA/RNA, proteins, lipids, and peptidoglycans1,2. In particular, the role of OMVs in bacterial pathogenesis has been extensively studied due to their enrichment in certain virulence factors and toxins3,4,5,6,7,8,9,10,11.
OMVs have been reported to range in size from 20 to 450 nm, depending on the parent bacteria and the growth stage, with several types of bacteria releasing heterogeneously sized OMVs8,12,13,14, which also differ in their protein composition and mechanism of host cell entry12. H. pylori released OMVs ranging in diameter from 20 to 450 nm, with the smaller OMVs containing a more homogeneous protein composition than the larger OMVs. Importantly, the two populations of OMVs were observed to be internalized by host cells via different mechanisms12. In addition, we have demonstrated that Aggregatibacter actinomycetemcomitans releases a population of small (<150 nm) OMVs along with a population of large (>350 nm) OMVs, with the OMVs containing a significant amount of a secreted protein toxin, leukotoxin (LtxA)15. While the role of OMV heterogeneity in cellular processes is clearly important, technical difficulties in separating and analyzing distinct populations of vesicles has limited these studies.
In addition to their importance in bacterial pathogenesis, OMVs have been proposed for use in a number of biotechnological applications, including as vaccines and drug delivery vehicles16,17,18,19,20. For their translational use in such approaches, a clean and monodisperse preparation of vesicles is required. Thus, effective and efficient methods of separation are necessary.
Most commonly, density gradient centrifugation (DGC) is used to separate heterogeneously sized vesicle populations from cellular debris, including flagellae and secreted proteins21; the method has also been reported as an approach to separate heterogeneously sized OMV subpopulations12,13,14. However, DGC is time-consuming, inefficient, and highly variable user-to-user22 and is, therefore, not ideal for scale-up. In contrast, size exclusion chromatography (SEC) represents a scalable, efficient, and consistent approach to purify OMVs21,23,24. We have found that a long (50-cm), gravity-flow, SEC column, filled with gel filtration medium is sufficient for efficiently purifying and separating subpopulations of OMVs. Specifically, we used this approach to separate A. actinomycetemcomitans OMVs into "large" and "small" subpopulations, as well as to remove protein and DNA contamination. Purification was completed in less than 4 h, and complete separation of the OMV subpopulations and removal of debris was accomplished.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. Preparation of buffers
- To prepare the ELISA wash buffer, add 3.94 g Tris-base, 8.77 g NaCl, and 1 g bovine serum albumin (BSA) to 1 L of deionized (DI) water. Add 500 µL polysorbate-20. Adjust the pH to 7.2 using HCl or NaOH.
- To prepare the blocking buffer, add 3.94 g Tris-base, 8.77 g NaCl, and 10 g BSA. Add 500 µL polysorbate-20 to 1 L of DI water. Adjust the pH to 7.2 using HCl or NaOH.
- To prepare the elution buffer (PBS), add 8.01 g NaCl, 2.7 g KCl, 1.42 g Na2HPO4, and 0.24 g KH2PO4 to 1 L DI water. Adjust the pH to 7.4 using HCl or NaOH.
NOTE: A 10x solution of this buffer can be made and diluted with DI water as needed.
2. Preparation of OMV sample
- Grow A. actinomycetemcomitans cells to the late exponential phase (optical density at 600 nm of 0.7). Pellet the cells by centrifuging twice at 10,000 x g at 4 °C for 10 min. Filter the supernatant through a 0.45 µm filter.
- Concentrate the bacteria-free supernatant using 50 kDa-molecular weight cut-off filters. Ultracentrifuge the concentrated solution at 105,000 x g at 4 °C for 30 min.
- Resuspend the pellet in PBS and ultracentrifuge again (105,000 x g at 4 °C for 30 min.) Resuspend the pellet in 2 mL of PBS.
3. Packing the S-1000 column
- Mix the stock bottle of gel filtration medium with a glass stir rod and pour out into a glass bottle the volume necessary to fill the column, plus approximately 50% excess (about 135 mL). Let these beads sit until they have settled, and then decant off the excess liquid. Resuspend the beads in elution buffer, so that the final solution is approximately 70% (by volume) gel, 30% buffer. Degas the solution under vacuum.
- Mount the glass column vertically using a ring stand and fill with elution buffer to wet the walls of the column. Drain the buffer until there is only about 1 cm of buffer remaining in the column.
- Without creating bubbles, carefully pipette beads into the column, filling the column to the top. Continue to drain excess buffer throughout this process. Be sure to not let the beads settle completely before adding additional beads to the top of the column. The column should be packed to a height of about 2 cm below the bottom of the column reservoir.
4. Loading the sample and collecting fractions
- Degas the elution buffer under vacuum. Wash the column with two column-volumes (180 mL) of elution buffer.
- Allow the remaining buffer to fully enter the column. Once the buffer has reached the top of the gel layer, carefully pipette a 2-mL sample containing OMVs (at a lipid concentration of approximately 100 - 200 nmol/L) onto the surface of the beads, being careful not to disturb any of the beads at the top of the column. Allow the sample to fully enter the gel, that is, when no liquid remains above the gel layer.
- Carefully and slowly add elution buffer on top of the gel column. Do not disturb the top layer of the gel, as this will cause sample dilution.
- Place a single 50-mL tube under the column and open the column. Collect the first 20 mL of the eluent. Add additional elution buffer to the top of the column, carefully, as needed to ensure the column is never dry.
- Place a series of 1.5 mL tubes under the column. Start the column and collect a series of 1-mL samples in each tube. As the samples are being collected, continue to add elution buffer to the top of the column, as necessary. Repeat until 96 fractions have been collected. Stop the column.
NOTE: The samples should be stored at -20 °C for long-term storage or 4 °C for short-term storage until further analysis. - To clean the column, run one column volume (90 mL) of 0.1 M NaOH through the column. Run two column volumes (180 mL) of elution buffer through the column.
5. Sample analysis
- To measure the lipid concentration in each fraction, pipette 50 µL of each fraction into a single well of a 96-well plate. To each well, add 2.5 µL of lipophilic dye. Incubate for 15 s. Measure the fluorescence intensity on a plate reader with an excitation wavelength of 515 nm and an emission wavelength of 640 nm. To calculate the fraction of all lipid in each sample, sum all of the emission intensities and divide each individual intensity by the total.
- To measure the concentration of a particular protein, pipette 100 µL of each fraction into a single well of an ELISA immuno-plate. Incubate at 25 °C for 3 h.
- Decant the samples. Add 200 µL of ELISA wash buffer to each well and decant. Repeat four times for a total of five washes.
- Add 200 µL of blocking buffer to each well and incubate for 1 h at 25 °C. Decant.
- Incubate plates with 100 µL blocking buffer plus primary antibody (1:10,000 for purified antibody; 1:10 for unpurified antibody) overnight at 4 °C. Decant.
- Add 200 µL of ELISA wash buffer to each well and decant. Repeat four times for a total of five washes.
- Add 100 µL of ELISA wash buffer plus secondary antibody (1:30,000) to each well. Incubate for 1 h at 25 °C.
- Add 200 µL of ELISA wash buffer to each well and decant. Repeat four times for a total of five washes.
- Add 100 µL of the 3,3',5,5'-tetramethylbenzidine (TMB) one-step solution and incubate for 15-30 min or until a blue color develops. Stop the TMB reaction with 50 µL of the stopping solution.
- On a plate reader, read the absorbance of each well at a wavelength of 450 nm.
- To measure the total protein concentration, record the absorbance at a wavelength of 280 nm (A280) of each fraction, using a UV-vis spectrophotometer.
A schematic of the protocol is shown in Figure 1.
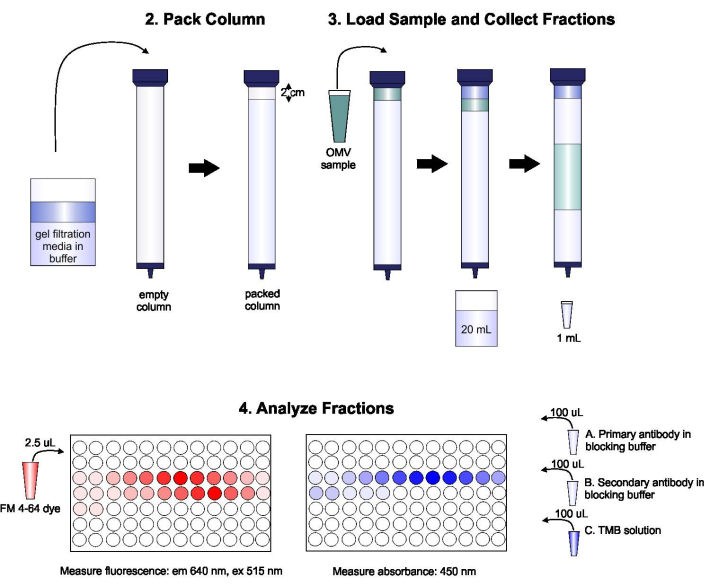
Figure 1: Schematic of SEC procedure. The column is packed with degassed gel filtration medium carefully to avoid bubbles and discontinuities, then washed with two column volumes of elution buffer. Next, the sample is carefully pipetted onto the top of the gel, without disrupting gel packing. The column is opened and run until the sample completely enters the gel. At this point, buffer is placed on the top of the column, and the first 20 mL of eluate is collected. Next, a series of 1-mL fractions is collected. These fractions are then placed in a 96-well plate or 96-well immuno-plate for analysis of lipid and protein content. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Results
Figure 2 shows representative results from this method. OMVs produced by A. actinomycetemcomitans strain JP2 were first purified from the culture supernatant using ultracentrifugation15. We previously found that this strain produces two populations of OMVs, one with diameters of about 300 nm and one with diameters of about 100 nm15. To separate these OMV populations, we purified the sample using the SEC protocol described above. E...
Access restricted. Please log in or start a trial to view this content.
Discussion
Here, we have provided a protocol for the simple, fast, and reproducible separation of bacterial OMV subpopulations. Although the technique is relatively straight-forward, there are some steps that must be performed extremely carefully to ensure that efficient separation occurs in the column. First, it is essential that the gel be loaded into the column carefully and slowly to avoid air bubbles. We have observed that leaving the gel at room temperature for several hours before loading the column allows the gel to equilib...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have no conflicts of interest to report.
Acknowledgements
This work was funded by the National Science Foundation (1554417) and National Institutes of Health (DE027769).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 1-Step Ultra TMB-ELISA | Thermo Scientific | 34028 | |
| Amicon 50 kDa filters | Millipore Sigma | UFC905024 | |
| Bovine Serum Albumin (BSA) | Fisher Scientific | BP9704-100 | |
| ELISA Immuno Plates | Thermo Scientific | 442404 | |
| FM 4-64 | Thermo Scientific | T13320 | 1.5 x 50 cm |
| Glass Econo-Column | BioRad | 7371552 | |
| Infinite 200 Pro Plate Reader | Tecan | ||
| Potassium Chloride (KCl) | Amresco (VWR) | 0395-500G | |
| Potassium Phosphate Monobasic Anhydrous (KH2PO4) | Amresco (VWR) | 0781-500G | |
| Sephacryl S-1000 Superfine | GE Healthcare | 17-0476-01 | |
| Sodium Chloride (NaCl) | Fisher Chemical | S271-3 | |
| Sodium Phosphate Dibasic Anhydrous (Na2HPO4) | Amresco (VWR) | 0404-500G | |
| Tris Base | VWR | 0497-1KG | |
| Tween(R) 20 | Acros Organics | 23336-2500 |
References
- Kuehn, M. J., Kesty, N. C. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes and Development. 19, 2645-2655 (2005).
- Kulp, A., Kuehn, M. J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annual Reviews Microbiology. 64, 163-184 (2010).
- Kato, S., Kowashi, Y., Demuth, D. R. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microbial Pathogenesis. 32 (1), 1-13 (2002).
- Nice, J. B., et al. Aggregatibacter actinomycetemcomitans leukotoxin is delivered to host cells in an LFA-1-independent manner when associated with outer membrane vesicles. Toxins. 10 (10), 414(2018).
- Haurat, M. F., et al. Selective sorting of cargo proteins into bacterial membrane vesicles. Journal of Biological Chemistry. 286 (2), 1269-1276 (2011).
- Horstman, A. L., Kuehn, M. J. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. The Journal of Biological Chemistry. 275 (17), 12489-12496 (2000).
- Wai, S. N., et al. Vesicle-mediated export and assembly of pore-forming oligomers of the Enterobacterial ClyA cytotoxin. Cell. 115, 25-35 (2003).
- Balsalobre, C., et al. Release of the Type I secreted α-haemolysin via outer membrane vesicles from Escherichia coli. Molecular Microbiology. 59 (1), 99-112 (2006).
- Donato, G. M., et al. Delivery of Bordetella pertussis adenylate cyclase toxin to target cells via outer membrane vesicles. FEBS Letters. 586, 459-465 (2012).
- Kim, Y. R., et al. Outer membrane vesicles of Vibrio vulnificus deliver cytolysin-hemolysin VvhA into epithelial cells to induce cytotoxicity. Biochemical and Biophysical Research Communications. 399, 607-612 (2010).
- Maldonado, R., Wei, R., Kachlany, S. C., Kazi, M., Balashova, N. V. Cytotoxic effects of Kingella kingae outer membrane vesicles on human cells. Microbial Pathogenesis. 51 (1-2), 22-30 (2011).
- Turner, L., et al. Helicobacter pylori outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Frontiers in Immunology. 9, 1466(2018).
- Zavan, L., Bitto, N. J., Johnston, E. L., Greening, D. W., Kaparakis-Liaskos, M. Helicobacter pylori growth stage determines the size, protein composition, and preferential cargo packaging of outer membrane vesicles. Proteomics. 19 (1-2), 1800209(2019).
- Rompikuntal, P. K., et al. Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infections and Immunity. 80 (1), 31-42 (2012).
- Nice, J. B., et al. Aggregatibacter actinomycetemcomitans leukotoxin is delivered to host cells in an LFA-1-independent manner when associated with outer membrane vesicles. Toxins. 10 (10), 414(2018).
- Stevenson, T. C., et al. Immunization with outer membrane vesicles displaying conserved surface polysaccharide antigen elicits broadly antimicrobial antibodies. Proceedings of the National Academy of Sciences. 115 (14), 3106-3115 (2018).
- Gujrati, V., et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 8 (2), 1525-1537 (2014).
- Huang, W., et al. Development of novel nanoantibiotics using an outer membrane vesicle-based drug efflux mechanism. Journal of Controlled Release. 317, 1-22 (2020).
- Chen, D. J., et al. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proceedings of the National Academy of Sciences. 107 (7), 3099-3104 (2010).
- Chen, L., et al. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proceedings of the National Academy of Sciences. 113 (26), 3609-3618 (2016).
- Singorenko, P. D., et al. Isolation of membrane vesicles from prokaryotes: A technical and biological comparison reveals heterogeneity. Journal of Extracellular Vesicles. 6 (1), 1324731(2017).
- Zeringer, E., Barta, T., Li, M., Vlassov, A. V. Strategies for isolation of exosomes. Cold Spring Harbor Protocols. 2015 (4), 319-323 (2015).
- Benedikter, B. J., et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Science Reports. 7 (1), 15297(2017).
- Mol, E. A., Goumans, M. J., Doevendans, P. A., Sluijter, J. P. G., Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine. 13 (6), 2061-2065 (2017).
- Lally, E. T., Golub, E. E., Kieba, I. R. Identification and immunological characterization of the domain of Actinobacillus actinomycetemcomitans leukotoxin that determines its specificity for human target cells. Journal of Biological Chemistry. 269 (49), 31289-31295 (1994).
- Chang, E. H., Giaquinto, P., Huang, J., Balashova, N. V., Brown, A. C. Epigallocatechin gallate inhibits leukotoxin release by Aggregatibacter actinomycetemcomitans by promoting association with the bacterial membrane. Molecular Oral Microbiology. 35 (1), 29-39 (2020).
- Klimentová, J., Stulík, J. Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiological Research. 170, 1-9 (2015).
- Dauros Singorenko, P., et al. Isolation of membrane vesicles from prokaryotes: a technical and biological comparison reveals heterogeneity. Journal of Extracellular Vesicles. 6 (1), 1324731(2017).
- Monguió-Tortajada, M., Gálvez-Montón, C., Bayes-Genis, A., Roura, S., Borràs, F. E. Extracellular vesicle isolation methods: rising impact of size-exclusion chromatography. Cellular and Molecular Life Sciences. 76 (12), 2369-2382 (2019).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved