Method Article
Analysis of Liver Microenvironment During Early Progression of Non-Alcoholic Fatty Liver Disease-Associated Hepatocellular Carcinoma in Zebrafish
In This Article
Summary
Here, we present how to generate a non-alcoholic fatty liver disease (NAFLD)-associated Hepatocellular Carcinoma (HCC) zebrafish model to study the impact of cholesterol surplus on liver microenvironment and immune cell landscape.
Abstract
Liver cancer is currently the third leading cause of cancer related death worldwide, and Hepatocellular Carcinoma (HCC) accounts for 75-90% of all liver cancer cases. With the introduction of effective treatments to prevent and treat hepatitis B/C, non-alcoholic fatty liver disease (NAFLD), and the more aggressive form know as non-alcoholic steatohepatitis (NASH), are quickly becoming the number one risk factors to develop HCC in modern societies. To better understand the role NASH has on the development of HCC we designed a NASH-associated HCC zebrafish. The optical clarity and genetic tractability of the zebrafish larvae make them an appealing and powerful model to study the liver microenvironment and immune cell composition using non-invasive fluorescent live imaging. This protocol describes how to use a NASH-associated HCC zebrafish model to investigate the effect of cholesterol surplus in the liver microenvironment and its impact on immune cell composition at early stages of the disease. First, we feed HCC larvae (s704Tg), which express hepatocyte-specific activated beta-catenin, with a 10% high cholesterol diet for 8 days to develop a NASH-associated HCC model. Here we describe how to make use of different transgenic lines to evaluate several early malignancy features in the liver by non-invasive confocal microscopy, such as liver area, cell, and nuclear morphology (hepatocytes area, nuclear area, nuclear:cytoplasmic ratio (N:C ratio), nuclear circularity, micronuclei/nuclear herniation scoring) and angiogenesis. Then, using transgenic lines with tagged immune cells (neutrophils, macrophages, and T cells) we show how to analyze liver immune cell composition in NASH-associated HCC larvae. The described techniques are useful to evaluate liver microenvironment and immune cell composition at early hepatocarcinogenesis stages, but they can also be modified to study such features in other liver disease models.
Introduction
Hepatocellular Carcinoma (HCC) is an aggressive cancer with limited therapeutic options. It has been found that upwards of 30% of all patients with HCC are obese and have NASH, an aggressive form of NAFLD1,2,3,4. Consumption of calorie rich diets drastically increase fatty acid availability that causes local and systemic metabolic shifts and triggers steatosis, hepatocyte injury, inflammation, and fibrosis - all key features of NASH. NASH progression to HCC involves the accumulation of lipids in the liver, which triggers inflammation and altered immune cell composition5,6,7. It is of particular interest and importance to understand how the liver microenvironment and immune cell landscape are altered during liver disease progression, and how it changes due to certain etiological factors. To better identify the impact that cholesterol surplus has on liver microenvironment and immune cell landscape, we have developed a unique zebrafish model of NASH-associated HCC. The use of this model has given us a better understanding of the impact of diet and overnutrition on liver microenvironment and liver disease progression.
Mammalian models, such as mice and human tissue samples, have been essential in understanding the pathogenesis of steatohepatitis and steatosis8. Mice are the preferred model for liver disease and cancer, but they lack optical clarity at a cellular level, while human tissue samples often lack the 3D environment that animal models are able to imitate. These obstacles have made Zebrafish a powerful model in the research community. Zebrafish have remarkable similarities to humans, with at least 70% gene conservation. They maintain liver microenvironment, hepatic cellular composition, function, signaling, and response to injuries9,10. Using a high-cholesterol diet (HCD) in combination with an established transgenic zebrafish model of HCC, we have developed a zebrafish model of NASH-associated HCC.
Here we present a protocol explaining how to generate a NASH-associated HCC zebrafish model and how to study liver microenvironment and address early malignancy features in vivo. Using non-invasive confocal microscopy in combination with zebrafish transgenic lines with fluorescently tagged hepatocyte membrane and nucleus, we can address early malignancy features by analyzing liver morphology (area, volume, and surface area), cell and nuclear morphology (hepatocytes area, nuclear area, N:C ratio, nuclear circularity, micronuclei/nuclear herniation scoring) and angiogenesis (vessel density). Immune cell microenvironment is also an important feature on hepatocarcinogenesis11,12,13,14, therefore, we also show how to analyze liver immune cell composition in NASH-associated HCC larvae, using transgenic zebrafish lines with tagged immune cells (neutrophils, macrophages, and T cells). The described techniques are unique to the model and extremely useful to evaluate liver microenvironment and immune cell composition in liver disease progression.
Protocol
Animal studies are carried out following procedures approved by the institutional animal care and use committee (IACUC) of Albert Einstein College of Medicine. For recipes for buffers and solutions used in the protocol please refer to Supplementary Table 1.
1. Preparation of 10% Cholesterol-enriched diet for acute cholesterol surplus.
- Weigh 2 x 4 g of Golden Pearl Diet 5-50 nm Active Spheres (GPAS) in two 25 mL glass beakers-one for the Normal Diet (ND) and the other one for the 10% High Cholesterol Diet (HCD).
NOTE: Any commercial dry food diet for zebrafish larvae can be used. - Weigh 0.4 g of Cholesterol in a 10 mL glass beaker. Cover the beaker with some foil.
- Measure 5 mL of Diethyl Ether using a 10 mL syringe with a 20 G needle. Next, add the Diethyl Ether to the ND beaker and immediately mix it with the dry food using a spatula.
CAUTION: Diethyl Ether causes eye, skin, and respiratory tract irritation. Use only in a chemical fume hood. - Measure again 5 mL of Diethyl Ether using a 10 mL syringe with a 20 G needle and add it to the 10 mL beaker with Cholesterol, mix immediately aspirating up and down with the syringe.
- Quickly add the Cholesterol solution to the HCD beaker and immediately mix it with a spatula until the solution is uniform.
- Leave beakers in the hood for up to 24 h to completely evaporate the Diethyl Ether. Next day, grind up GPAS diets into fine particles using a pestle and mortar.
NOTE: Grinding the diets is a crucial step, larvae will not be able to eat particles bigger than 50 nm. - Transfer each diet to a small, labeled plastic bag or to a 50 mL centrifuge tube and store at -20 ˚C.
2. NASH induction with short-term larvae feeding with cholesterol-enriched diet - static conditions.
- Set up transgenic fish lines on Day -1 (Table 1). The next morning (Day 0), after fish have spawned, collect eggs in a mesh strainer.
- Using a wash bottle with embryo water (E3), rinse eggs thoroughly and carefully transfer the eggs to a 10 cm Petri dishes with E3 medium.
- Clean the plates of all debris that might have accumulated in the breeding boxes, including any dead or unfertilized eggs.
NOTE: Rinsing the eggs and cleaning of the plates is crucial to avoid uncontrolled growing of microorganisms and consequent defects on larvae development. - Divide eggs in a density of 70/80 per 10 cm petri dishes (in 25 mL of E3). At Day 1, under a dissection scope equipped with a transillumination base, check and clean dead embryos or embryos with developmental defects.
NOTE: Use transillumination base darkfield mode to identify embryos with development effects and to check for growth of microorganisms in the plates. Different facilities have different microorganism yield in their systems. Change E3 medium and/or dishes if necessary, to avoid negative effects. - At Day 5, combine larvae from all the dishes in a 15 cm Petri dish, add E3 without methylene blue. Next, divide larvae in the feeding boxes as following (Table 2).
NOTE: In control conditions, to generate healthy larvae without triggering systemic chronic inflammation, larvae should be fed with around 0.1 mg of food per larvae per day. Notice that total amount of food (Table 2) is divided equally in two feedings per day. - Keep larvae in an incubator at 28 °C, in a dark-light cycle (e.g., 14 h light/10 h dark cycle), and feed larvae twice a day from Day 5 to Day 12.
- Aspirate daily any food debris, as well as 90-95% of the medium using a vacuum system attached to a 1 mL pipette tip. Next carefully pour new E3 without methylene blue to one corner of the feeding box to avoid damaging the larvae.
NOTE: Daily cleaning of the feeding boxes is crucial to avoid the uncontrolled growth of microorganisms.
3. Collection of 13 days post fertilization larvae from feeding boxes.
- At Day 13, prepare collection dishes accordingly to the number of experimental conditions that were set up. With a permanent marker pen make a mark on the side of the dishes (lid and bottom) to avoid switching samples, for example one black stripe for normal diet (ND) and two black stripes for HCD.
- Pour enough E3 without methylene blue to cover the bottom of each dish. Carefully using the vacuum system aspirate the water from the feeding boxes, try to aspirate any debris or dead larvae from the bottom to avoid transferring these materials to the collection dishes.
- When water level starts to get low, slowly lift the feeding box to make larvae swim to one of the corners, then aspirate in the opposite direction.
NOTE: Use 1 mL pipette with cut tips, at different heights, to allow for different diameters when aspirating the water. Alternate between the different sizes, start with a larger diameter, and change to a smaller one as the water volume reduces and larvae density increases. - Once there is only about 20-30 mL of liquid, carefully decant larvae into the collection petri dish prepared in Step 1. Place the labeled tape from the feeding box on the lid of the dish.
- Repeat steps 3.2-3.4 for all the feeding boxes.
4. Control assay for diet induced hepatic steatosis - Oil Red O (ORO) staining, imaging, and scoring.
- Using a glass Pasteur pipette, transfer 15-20 larvae to a 2 mL round bottom tube and place on ice for about 15-30 min. Remove most of the media from the tube.
- Add 2 mL of 4% of paraformaldehyde (PFA) to fix larvae and incubate at 4 °C overnight.
- Next morning, remove 4% PFA solution and wash larvae three times with 2 mL of 1x PBS.
- Prepare a 12 well plate (Figure 2A) per condition with one mesh well insert.
- Using a glass pipette, transfer larva from the 2 mL tube to the insert previously placed in the well, add 5 mL of 1x PBS. Save the 2 mL tubes for sample storage after staining.
CAUTION: Larvae are extremely sticky at this stage do not use plastic pipettes. - Transfer insert with larvae to another well with 5 mL of 60% Isopropanol solution. Incubate larvae for 30 min room temperature (RT).
- Meanwhile, prepare a 0.3% Oil Red O in 60% Isopropanol solution. Filter the 0.3% Oil Red solution twice using a 0.45 µm syringe filter.
NOTE: Prepare 5 mL of solution per well by mixing 3 mL of 0.5% Oil Red O in Isopropanol with 2 mL of distilled water. The 0.3% Oil Red O in 60% Isopropanol solution needs to be freshly prepared. - Next, transfer the mesh insert with larvae to another well with 5 mL of freshly made 0.3% Oil Red O in 60% Isopropanol solution. Incubated larvae for 3 h at RT with gentle rocking.
- After ORO staining, rinse larvae by transferring insert to another well with 60% Isopropanol. Wash larvae twice in 60% Isopropanol for 30 min each time by transferring insert sequentially to wells with 60% Isopropanol.
- Wash larvae three times for 5 min in 1x PBS-0.1% Tween by transferring insert sequentially to wells with 1x PBS-0.1% Tween.
- Transfer larvae to the 2 mL tubes. Remove PBST, add 1 mL of 80% Glycerol, and store at 4 °C until imaging.
- For better imaging, transfer larvae to 1x PBS-0.1% Tween and under a dissection scope dissect livers by carefully pulling tissues using two #55 forceps.
NOTE: If using a Casper background imaging can be performed using the whole larvae. - Using a glass pipette, carefully transfer livers to a well of a Porcelain Spot Plate with 50% Glycerol. Position livers using a small tool to manipulate larvae (e.g., eyelash tool- Figure 2B). Image livers in a stereomicroscope equipped with a color camera.
- Score hepatic steatosis (no ORO staining = none; light red ORO staining = mild; red-dark red ORO staining = moderate/severe).
5. Non-invasive confocal imaging using the zebrafish Wounding and Entrapment Device for Growth and Imaging(zWEDGI).
- Prepare a 10 cm Petri dish with 15-20 mL of 1x Tricaine-E3, just enough to cover the bottom.
NOTE: A 1x Tricaine-E3 working solution (0.16 mg/mL) can be obtained by diluting 2 mL of a 25x Tricaine stock solution (4 mg/mL) in 48 mL of E3. Tricaine concentration must not exceed 0.16 mg/mL since larvae are extremely sensitive to the anesthesia at this development stage. - Transfer 15-20 larvae with a plastic Pasteur pipette to the Petri dish containing 1x Tricaine-E3 and anesthetize larvae for 5 min.
- Prepare a 6 well plate with 2-3 mL of E3 without methylene blue per well.
- On a fluorescent stereomicroscope, screen the anesthetized larvae for desired fluorescent markers (Table 1). Transfer screened larvae in the minimum amount of Tricaine in E3 and let them to recover in the 6 well plate until it is time to image.
NOTE: Screening of double, triple, and quadruple transgenic larvae takes time, place larvae in E3 and change media from the 6 well plate often to decrease unnecessary exposure to Tricaine. Also, larvae at this developmental stage float so use an eyelash tool to position larvae for screening without inducing tissue damage. - After screening, prepare a 10 cm Petri dish with 25 mL of 1x Tricaine-E3. Transfer 15-20 larvae with a plastic Pasteur pipette to the petri dish containing 1x Tricaine-E3 and anesthetize larvae for 5 min.
- Meanwhile, under the stereomicroscope add 1x Tricaine-E3 into the chambers of the wounding and entrapment device (e.g., zWEDGI)15,16. Remove air bubbles from the chambers and the restraining tunnel using a P200 micropipette. Remove all excess of 1x Tricaine-E3 leaving only enough volume to fill the chambers.
- Transfer an anesthetized larva into the loading chamber of the zWEDGI, and position it using an eyelash tool under the stereomicroscope. Gently, tap larvae's head and push it so that the tail enters the restraining tunnel. In addition, using the micropipette remove 1x Tricaine-E3 from the wounding chamber to help larva enter the tunnel. Ensure that the larva is positioned appropriately for imaging of the left lobe - Inverted Microscope: left side facing down; Upright Microscope: left side facing up.
NOTE: If a zWEDGI is not available, fish can be mounted and positioned in 1% low melting point agarose in 1x Tricaine-E3, however, agarose immersion can be used just for quick imaging (up to 15 min). - For hepatocyte and nuclear morphology analysis, image livers with a confocal microscope using a 40x air objective and z-stacks of 2 µm optical sections. For liver morphology, angiogenesis, and immune cell recruitment assays image livers using a 20x air objective and z-stacks of 5 µm optical sections. For larvae with large livers, 2 x 2 tile images should be taken to assure imaging of all liver and surrounding area of 75 µm.
- After imaging, use a plastic Pasteur pipette with 1x Tricaine-E3 to pull larva from the restraining tunnel and transfer to a petri dish with E3 without methylene blue.
6. Analysis of liver morphological variations.
NOTE: The steps listed below are performed for liver surface area and volume quantification:
- Open imaging software. Add folder containing image files to be analyzed to Arena selecting Observe folder icon. Next, by right clicking on thumbnails select Convert to native Imaris file format to convert files into the format (.ims).
- On Surpass submenu, adjust brightness and contrast for each channel. Next, create a surface from liver clicking on the blue Add new surfaces button.
- Next, on surface creation panel select Segment only a region of interest to manually create a surface of the liver.
- Click Skip automatic creation | Edit manually option. Select Contour and de-select "Volume" option at the scene panel.
NOTE: Volume view option needs to be unselected, otherwise selection of region of interest in different z-stacks will not be able. - Navigate through z-stacks and draw region of interest in each plane by selecting Mode, pick the drawing mode that you prefer. To start drawing change pointer to Select mode and not Navigate, next click on Draw and start dragging the pointer thru liver to draw a ROI.
- After selecting a ROI in the different focal planes select Create a surface to merge all the selected ROIs and create liver surface.
- Next, using the newly created surface create a mask of the hepatocytes signal. Click on the surface and under the Edit tab (the pencil) choose Mask All. In the window that appears, choose the channel you wish to mask that will be used to quantify liver volume and surface area. Then, choose set voxels outside surface to 0 and tick on the Duplicate channel option before applying mask.
- Use liver surface area and liver volume values from statistical analysis menu for analysis.
NOTE: Perform the following steps for liver area quantification.
- Open Fiji software. On Plugins menu, select Bio-formats followed by Bio-formats Importer, tick on Split channels option to open image file.
- On Image menu, select Properties to check that image has the correct pixel size and voxel depth, if not correct values. Next, go to Images Menu, click on Adjust followed by Brightness and Contrast, adjust image brightness and contrast.
- Next, using hepatocytes channel create a maximum intensity projection of the liver. Go to Images Menu, click on Stacks followed by Z Project. Select Maximum Intensity Projection.
- Using the freehand selection tool, manually create a region of interested (ROI) surrounding the larval liver. Next, on Analyze menu click on Measure to obtain liver area. Save results as spreadsheet for further analysis.
7. Hepatocyte and Nuclear morphological analysis.
- Open Fiji software. On Plugins menu, select Bio-formats followed by Bio-formats Importer tick on Split channels option to open image file.
- On Image menu, select Properties to check that image has the correct pixel size and voxel depth, if not correct values. Next, go to Images menu, click on Adjust followed by Brightness and Contrast, adjust image brightness and contrast.
- In the Analyze menu select Set Measurements tick on all the parameters that you want to analyze (e.g., area, perimeter, and shape descriptors). Select hepatocyte membrane fluorescent channel. Navigating through Z, use the freehand selection tool to draw perfect ROI around one hepatocyte. Next, on Analyze menu click on Measure to calculate hepatocytes area.
- Repeat step 7.3 for 15-30 hepatocytes. Save results as spreadsheet for further analysis.
- Next, select nucleus fluorescent channel. Navigating through Z, use the freehand selection tool to draw perfect ROI around hepatocyte nucleus. Next, on Analyze menu click on Measure to calculate nuclear area and circularity.
- Repeat step 7.5 for 15-30 hepatocytes. Save results as a spreadsheet for further analysis, including quantification of nuclear:cytoplasmic ratio.
- Score samples for presence of micronuclei and herniation as following (Table 3).
8. Angiogenesis analysis.
- Manually create a surface for liver as described on section 6 of this protocol. Name this surface as "Liver Surface".
- Create a mask for the endothelial cells signal inside the liver. Click on the surface and under the edit tab (the pencil) choose Mask All. In the window that appears, choose the endothelial cells channel to isolate vessel signals just from the liver. Choose set voxels outside surface to 0 and tick on the Duplicate channel option before applying Mask.
- Rename new endothelial cells masked channel as Liver Vasculature.
- To measure vessel volume and surface area create a second surface. Click on the blue Add new surfaces button. In the first step of the surface creation make sure all options are unselected to automatically create a surface.
- In the second step choose Liver Vasculature channel to create a surface over. Deselect smoothing option to detect as many details from vessels as possible and enable background subtraction in the thresholding option.
- Adjust thresholding assuring that detected surface is colocalizing with liver vasculature signal. Use surface area and volume values from statistical analysis menu. Calculate vessel density index using the following equations:
Vessel density index (by Liver µm2) = (Vessel Surface Area (µm2)) / (Liver Surface Area (µm2))
Vessel density index (by Liver µm3) = (Vessel Volume (µm3)) / (Liver Volume (µm3))
9. Immune cell recruitment analysis.
- Open Fiji software. On Plugins menu, select Bio-formats followed by Bio-formats Importer tick on Split channels option to open image file.
- On Image menu, select Properties to check that image has the correct pixel size and voxel depth, if not correct values. Next, go to Images menu, click on Adjust followed by Brightness and Contrast, adjust image brightness and contrast (e.g., macrophages - mCherry or dTomato; neutrophils - BFP; T-cells - EGFP; hepatocytes - EGFP or mCherry).
- Next, create maximum intensity projections to each channel.
- As described in section 6 (Steps 6.9-6.12), create a ROI surrounding the larval liver and measure liver area. Create a second ROI including liver area and 75 µm surrounding area ("Recruitment area").
- Next, on Edit menu select Selection option followed by Add to manager. Selecting an innate immune cell channel Maximum Intensity Projection, click on the liver ROI from the ROI Manager window to set the Recruitment area.
- On Plugins menu, select Analyze option followed by Cell Counter and count immune cells inside the Recruitment area. Record number of recruited immune cells to the liver area on an spreadsheet.
- Calculate neutrophil, macrophage, and T-cell densities by normalizing the number of immune cells per liver area.
Results
By introducing a short-term high cholesterol diet into a Hepatocellular Carcinoma (HCC) zebrafish model, which overexpresses a hepatocyte specific constitutively active form of Beta-catenin (s704Tg; Tg(fabp10a:pt-B-cat, cryaa:Venus)17, we are able to create a non-mammalian vertebrate model of NASH-associated HCC. Liver disease progression can be monitored early by measuring hepatic steatosis, liver size, hepatocyte, nuclear morphology, angiogenesis, and immune cell infiltration (Figure 1).
HCC larvae fed with a normal diet show none to mild hepatic steatosis, measured by Oil Red O staining. However, HCC larvae fed with a high cholesterol diet show a significant increase in hepatic steatosis (Figure 2).
A well-known marker of liver disease is hepatomegaly17. To evaluate liver size, HCC larvae can be outcrossed with a transgenic line specifically expressing a fluorescent marker on hepatocytes, such as the Tg(fabp10a:H2BmCherry). To assess hepatomegaly, evaluation of the liver area (2D), liver surface area, and liver volume (3D) are performed. After 8 days of exposure to a cholesterol surplus, liver enlargement was observed in in HCC larvae (Figure 3).
Using non-invasive live imaging, transgenic fish lines expressing fluorescent proteins in hepatocyte membranes (such as Tg(fabp10a:Life-actin-EGFPP) and in hepatocytes nuclei (such as Tg(Fabp10a:H2B-mCherry) can be used to assess cellular and nuclear morphology alterations associated with malignancy in hepatocytes. Hepatocyte area was increased in NASH-associated HCC (Figure 4A,B,D), as well as nuclear area (Figure 4C,E) and nuclear:cytoplasmic ratio (Figure 4F). A significant decrease in nuclear circularity was also observed in the HCD+HCC group (Figure 4G). Lipotoxicity triggers DNA damage, a feature of carcinogenesis in the presence of micronuclei. Using the H2B-mCherry marker we observed a greater incidence of micronuclei in the HCC larvae fed with a high cholesterol diet (Figure 4H).
Hepatic vasculature can easily be evaluated in zebrafish models using a transgenic tagged line, such as Tg (kdrl:mCherry or Tg(fli:EGFP), which label vasculature. A significant increase of vessel density was observed in HCC+HCD larvae (Figure 5).
To observe the inflammatory response triggered early in NASH-associated HCC, a transgenic fish line expressing fluorescent proteins in macrophages and neutrophils, such as Tg(mfap4:tdTomato-CAAX; lyz:BFP), was outcrossed with the HCC transgenic line. Infiltration of neutrophils and macrophages occur in both HCC and HCC fed with HCD, which was assessed by quantification of number and density of neutrophils/macrophages in the liver and vicinity (surrounding area up to 75µm) (Figure 6A-H). Nevertheless, the HCC+HCD displayed a significant increase in neutrophil number and density (Figures 6F-H). At 13 days post fertilization, the adaptive immune system is already functional. Using a transgenic fish line expressing fluorescent protein in T-Cells, such as the Tg(lck:EGFP), and in combination with the HCC transgenic line; we evaluated the impact of cholesterol surplus on recruitment of T-cells to the liver. A significant decrease in T-Cell density and overall number was observed in HCC larvae fed with HCD (Figure 6I-K).
| Transgenic Zebrafish lines | ZFIN reference | Assay |
| Casper | roya9; mitfaw2 | Hepatic steatosis |
| Tg(fabp10a:pt-β-catenin_cryaa:Venus / fabp10a:H2B-mCherry ) | s704Tg / uwm41Tg | Liver morphology |
| Tg(fabp10a:pt-β-catenin_cryaa:Venus; fabp10a:H2B-mCherry; fabp10a:LIFEACT-EGFP) | s704Tg / uwm41Tg / uwm42Tg | Hepatocytes and nuclear morphology |
| Tg(fabp10a:pt-β-catenin_ cryaa:Venus; fli:EGFP) | s704Tg / y1Tg | Angiogenesis |
| Tg(fabp10a:pt-β-catenin_cryaa:Venus; mfap4:Tomato-CAAX; lyzC:BFP) | s704Tg / xt6Tg / zf2171Tg | Macrophage and Neutrophil recruitment |
| Tg(fabp10a:pt-β-catenin_cryaa:Venus; lck:EGFP) | s704Tg / cz1Tg | T-cells recruitment |
Table 1: Transgenic zebrafish lines to use in different assays.
| Number of larvae | Feeding box size | Amount of food per day (mg) | E3 Volume (ml) |
| 30-40 | Small breeding box | 3-4 | 200 |
| 60-80 | Small/Big breeding box | 6-8 | 400 |
| 100-150 | Big breeding box | 10-15 | 500 |
Table 2: Set up conditions of feeding boxes.
| Phenotype | Scoring method |
| None | Normal Nuclei, No micronuclei or herniation |
| Mild | Low number of micronuclei (less than 5 per field of view) and/or herniation |
| Moderate/ Severe | Moderate to high number of micronuclei (more than 5 per field of view) and/or herniation |
Table 3: Micronuclei and Nuclear Herniation Scoring.
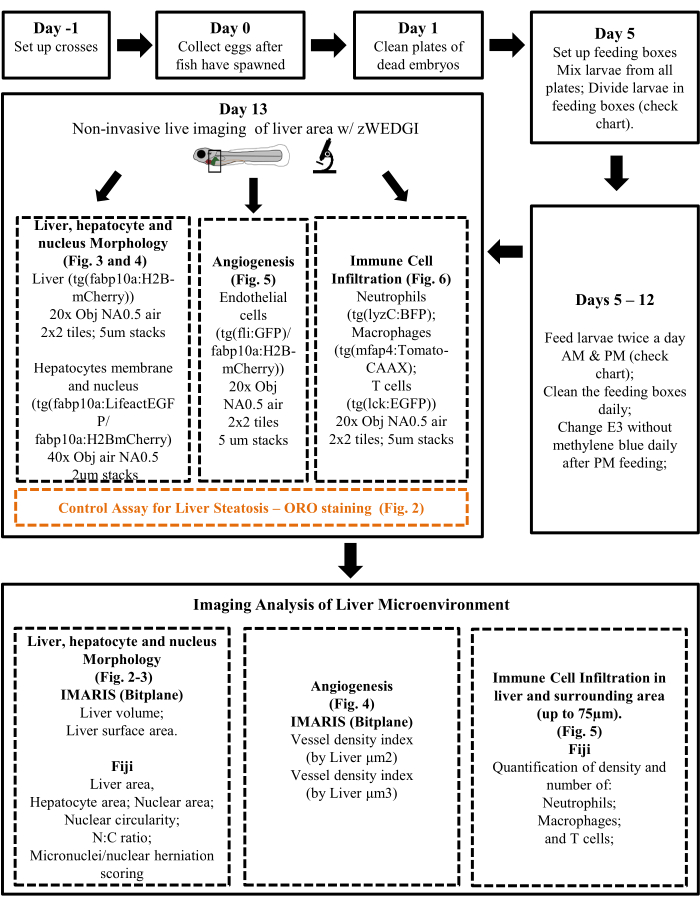
Figure 1: Protocol Diagram summarizing main experimental steps and analysis approach. Please click here to view a larger version of this figure.
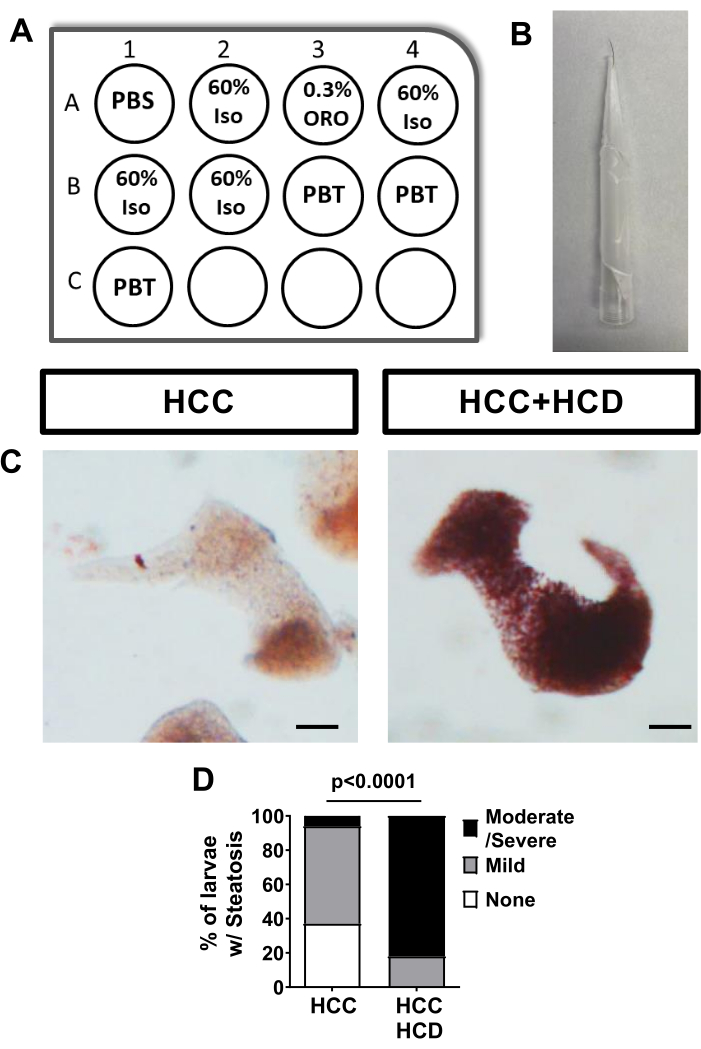
Figure 2: Control Assay for Liver Steatosis - ORO staining. HCC larvae were fed with normal or high cholesterol diet and Oil Red O (ORO) staining was performed to assess hepatic steatosis. (A) Diagram of the 12 well plate to perform sequential ORO staining using the mesh well inserts. (B) Image of eyelash tool used to manipulate larvae. (C) Representative images of livers stained with Oil Red; HCC and HCC+HCD larvae. (D) Chi-square graph showing percentage of larvae with different scoring of liver steatosis. Scale bar= 50 µm. Please click here to view a larger version of this figure.
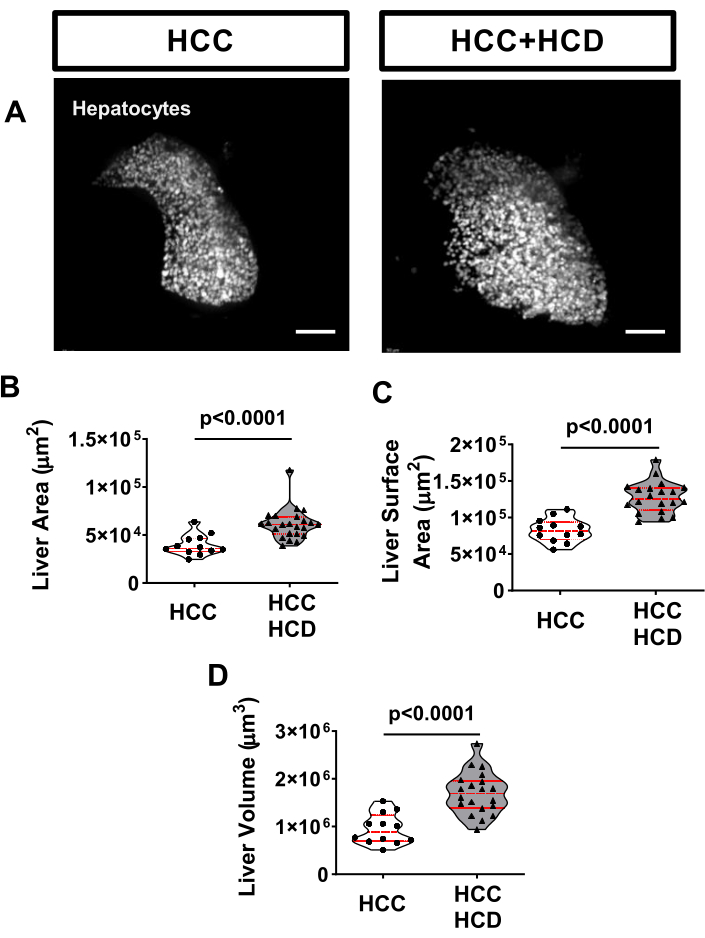
Figure 3: Representative images from liver size. Transgenic HCC larvae expressing a liver marker (Tg(fabp10a:H2B-mCherry)) were imaged live and non-invasively, on an inverted spinning disk confocal microscope using a zWEDGI. (A) Representative 3D reconstructions of livers in HCC and HCC+HCD larvae. (B-D) Graph showing liver morphological alterations including liver area (B) liver surface area (C) and liver volume (D) in HCC and HCC+HCD larvae. Scale bar = 50 µm. Please click here to view a larger version of this figure.
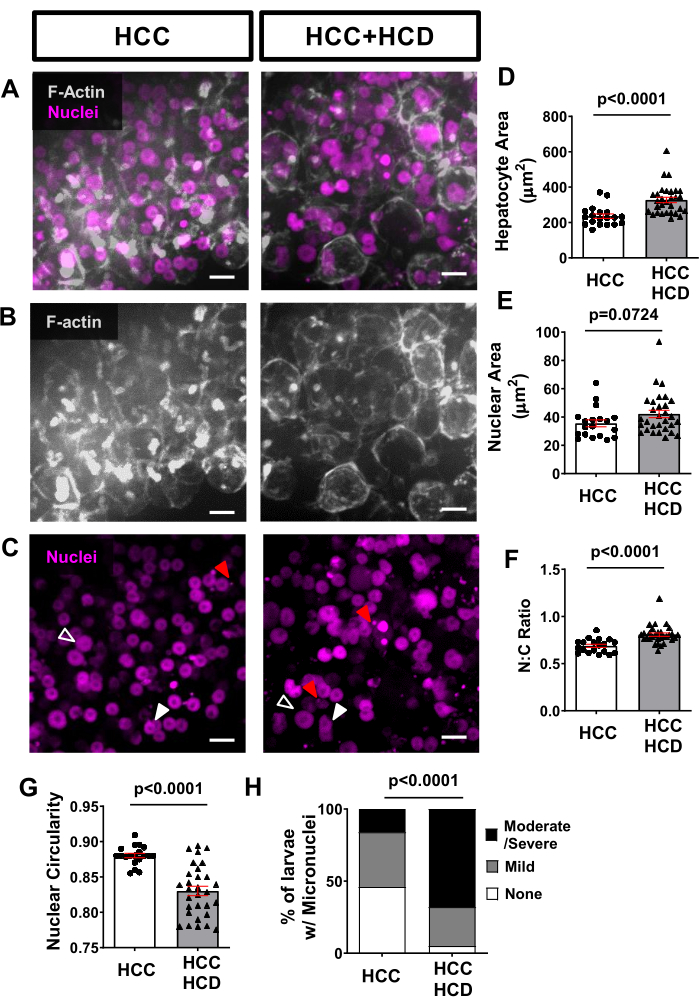
Figure 4: Representative images from hepatocytes and nuclei morphology. Transgenic HCC lines expressing fluorescent proteins in hepatocyte membranes (Tg(fabp10a:Life-actin-EGFP) and in hepatocytes nuclei (Tg(fabp10a:H2B-mCherry) were imaged. (A-C) Representative 3D reconstructions of F-actin and hepatocyte nuclei of HCC and HCC+HCD larvae. Open arrowheads show enlarged nuclei; white arrowheads show nucleus with altered shape; and red arrows show micronuclei and nuclear herniation. (D-G) Graphs showing averages of cell and nuclear parameters in HCC and HCC+HCD 13-day old larvae. (D) Hepatocytes area. (E) Nuclear Area. (F) Nuclear:Cytoplasm ratio. (G) Nuclear circularity. Each dot represents averages per larva. Dot plots show mean ±SEM (H) Chi-square graphs showing percentage of larvae with different scoring of Micronuclei and Nuclear herniation. Scale bar = 10 µm. Please click here to view a larger version of this figure.
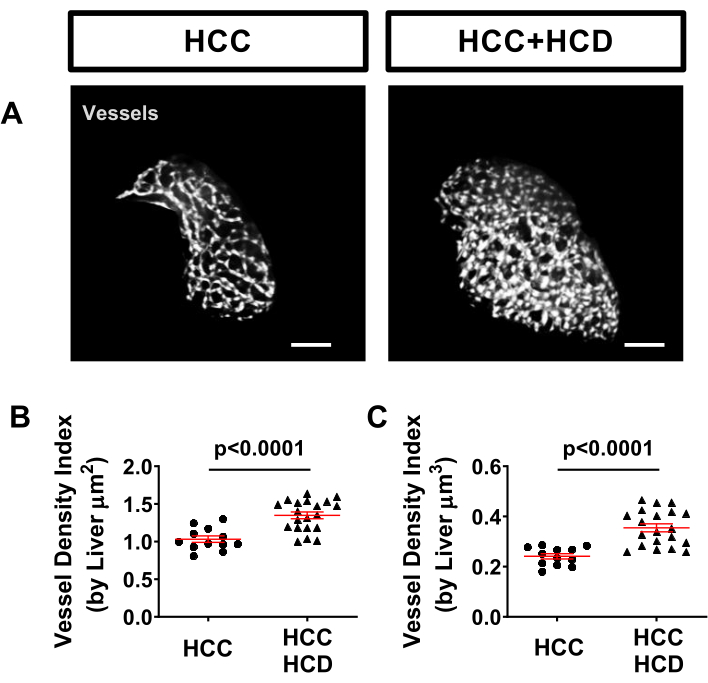
Figure 5: Representative images from hepatic vasculature. Transgenic HCC lines expressing fluorescent proteins in hepatocyte nuclei (Tg(Fabp10a:H2B-mCherry) and in endothelial cells (Tg)fli:EGFP)). (A) Representative 3D reconstructions of hepatic vasculature in HCC and HCC+HCD larvae. (B-C) Graph showing vessel density index by liver surface area (B) volume (C) in HCC and HCC+HFCD larvae. Dot plots show mean ±SEM. Scale bar = 50 µm. Please click here to view a larger version of this figure.
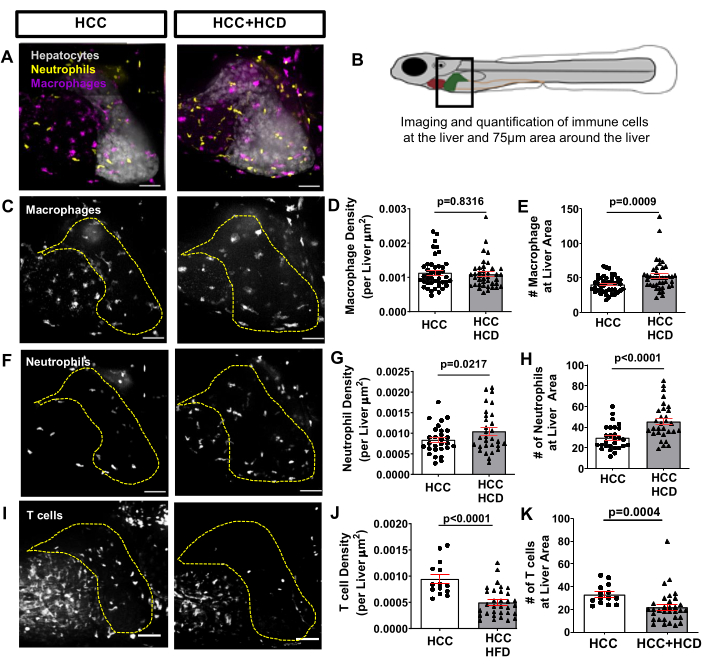
Figure 6: Representative images from liver immune cell landscape. Transgenic HCC line expressing fluorescent proteins in macrophages and neutrophils (Tg(mfap4:tdTomato-CAAX; lyz:BFP) or in T-cells (Tg(lck-EGFP). (A,C,F) Representative 3D reconstructions of livers and leukocyte recruitment to liver area in 13-day old HCC and HCC+HCD larvae. (B) Diagram of imaged liver area. (D-E) Graph showing macrophage density (D) and number (E) in liver area in HCC and HCC+HCD larvae. (G-H) Graph showing neutrophils density (G) and number (H) in liver area in HCC and HCC+HCD larvae. (G) Representative 3D reconstructions of T cell recruitment to liver area in HCC and HCC+HCD larvae. (H-I) Graph showing T cell density (H) and number (I) in liver area in HCC and HCC+HCD larvae. Dot plots show mean ±SEM. Scale bar = 50 µm. Please click here to view a larger version of this figure.
Supplementary Table 1: Table of Buffers and solutions Please click here to download this Table.
Discussion
With an increased incidence of HCC, specifically NASH induced HCC, it is of great importance to have more efficient models to study the cellular and molecular mechanisms involved in NASH-associated HCC. Deconvolution of cell-cell interactions at the liver are crucial to better understand liver disease progression and hepatocarcinogenesis. The approach described in this protocol offers a unique way to analyze liver disease progression in vivo and non-invasively.
Preparation of diet is critical to the success of establishing a NASH-associated HCC model. It is important to let diethyl ether completely evaporate inside the fume hood to avoid harmful effects, while preparing diets for zebrafish. To use these diets with larvae (5-12 days post fertilization), it is extremely important to grind up the diets into fine particles to assure the food intake by the larvae. The use of fluorescently tagged fatty acid analogs can be used to evaluate food intake in larvae.
Before placing the larvae into the breeding boxes and starting the feeding procedure, it is crucial to ensure that experimental sampling is uniformized by mixing larvae from different plates. This step is important since different microenvironments, beginning at Day 0, are promoted in each of the plates and might affect inflammatory response.
Another important step is to count the larvae, to know how much food is needed for each feeding box. If the amount of food is not adequate for the number of larvae present in each box, one of the two scenarios will occur: 1) larvae will be underfed; 2) larvae will be overfed. Inaccurate feeding will lead to unhealthy conditions associated with malnutrition or overnutrition, such as inflammation, which will drastically affect the liver microenvironment. If inaccurate feeding occurs, larvae will show motion issues. At this development stage, larvae should be swimming intensively, therefore if motion issues are noticed larvae were raised under unhealthy conditions (malnutrition or exposure to toxic doses of cholesterol due to overfeeding). For this reason, feeding procedures need to be tightly controlled for both diets, normal and cholesterol enriched. Some assays can be performed to quickly address accuracy of the feeding and health of larvae including presence of hepatomegaly (liver size) and inflammation of tissue and organs, particularly liver and intestine (visible increased infiltration of neutrophils and macrophages).
Daily cleaning and replacement of 95% of E3 is pivotal to reduce the growth of microorganisms in the feeding boxes and improve larvae health and survival. Alternatively, larvae can be placed in a Stand-alone Rack System. For best results, place 60-80 larvae in a 3-liter tank. Keep water flow at the minimum level by adjusting the flow to a fast-dripping mode and feeding larvae 3-4 mg twice a day (AM and PM). Water flow should be checked regularly to assure correct flow in each tank. In our laboratory, this method gives us 95-100% survival with short-term feeding of 10% HCD. In addition, this method greatly reduced the workload inherent to the daily cleaning and water exchange necessary in the static feeding protocol described.
While we utilized a 10% cholesterol-enriched diet to induce NASH in a short-term exposure (5 days is enough to induce steatohepatitis), diet alterations can be performed and expanded to use fructose 18, fatty acids (such as Palmitic Acid)19, or feeding protocols that can be extended using a 4% cholesterol-enriched diet20. Currently, there are few successful therapeutic targets for HCC and none for NASH. The use of zebrafish models offers a unique opportunity to expand our knowledge on hepatocarcinogenesis but also an unparallel vertebrate system to perform large throughput drug screenings. The techniques described in this protocol will facilitate future findings and therapeutic targets for liver disease and hepatocarcinogenesis.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The author would like to acknowledge the Albert Einstein College of Medicine Zebrafish Core Facility technicians Clinton DePaolo, and Spartak Kalinin for assistance and maintenance of our zebrafish lines. FJMN is supported by the Cancer Research Institute and Fibrolamellar Cancer Foundation.
Materials
| Name | Company | Catalog Number | Comments |
| Cholesterol | Sigma | C8667-25G | Easily degraded. Store -20°C. |
| Corning Netwells carrier kit 15 mm | Fisher | 07-200-223 | |
| Corning Netwells inserts | Fisher | 07-200-212 | |
| Diethyl ether | Fisher | 60-046-380 | Highly Volatile. |
| Dumont forceps #55 dumostar | Fisher | NC9504088 | |
| Fisherbrand Pasteur Pipets 5.75in | Fisher | 22-183624 | |
| 4% paraformaldehyde (PFA) | Electron Microscopy Science | 15710 | |
| Golden Pearl Diet 5–50 nm Active Spheres | Brine Shrimp Direct | - | Any commercial dry powder food for larvae can be used. |
| Graduated Transfer Pipets | Fisher | 22-170-404 | |
| Isopropanol | Fisher | BP26181 | |
| PBS, pH7.4, 10X, 10 Pack | Crystalgen | 221-1422-10 | |
| Petri Dishes 100X20MM | Fisher | 08-747D | |
| Tricaine | Sigma | A-5040 | |
| Tween 20 | Fisher | BP337-500 | |
| Oil Red O solution 0.5% isopropanol | Sigma | O1391-500ML | |
| Tricaine | Sigma | A-5040 | |
| Tween 20 | Fisher | BP337-500 | |
| Vactrap | VWR | 76207-630 | Vacuum system for larvae collection |
| Microscopes | |||
| Fluorescent Stereomicroscope | Leica | M205 FCA THUNDER Imager Model Organism Large | |
| Spinning Disk Confocal Microscope | Nikon | Nikon CSU-W1 | |
| Stereomicroscope | Leica | S9i with transilluminated base | |
| Software | |||
| Fiji | Open-source Java image processing program. | ||
| Imaris 9.6 | Bitplane; Oxford Instruments. |
References
- Pocha, C., Kolly, P., Dufour, J. F. Nonalcoholic Fatty Liver Disease-Related Hepatocellular Carcinoma: A Problem of Growing Magnitude. Seminals in Liver Disease. 35 (3), 304-317 (2015).
- El-Serag, H. B., Kanwal, F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go. Hepatology. 60 (5), 1767-1775 (2014).
- Estes, C., et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016-2030. Journal of Hepatology. 69 (4), 896-904 (2018).
- Estes, C., Razavi, H., Loomba, R., Younossi, Z., Sanyal, A. J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 67 (1), 123-133 (2018).
- Meli, R., Mattace Raso, G., Calignano, A. Role of innate immune response in non-alcoholic Fatty liver disease: metabolic complications and therapeutic tools. Frontiers in Immunology. 5, 177 (2014).
- Ganz, M., et al. Progression of non-alcoholic steatosis to steatohepatitis and fibrosis parallels cumulative accumulation of danger signals that promote inflammation and liver tumors in a high fat-cholesterol-sugar diet model in mice. Journal of Translational Medicine. 13, 193 (2015).
- Ma, C., et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 531 (7593), 253-257 (2016).
- Reimer, K. C., Wree, A., Roderburg, C., Tacke, F. New drugs for NAFLD: lessons from basic models to the clinic. Hepatology International. 14 (1), 8-23 (2020).
- Wrighton, P. J., Oderberg, I. M., Goessling, W. There is something fishy about liver cancer: Zebrafish models of hepatocellular carcinoma. Cellular and Molecular Gastroenterology and Hepatology. 8 (3), 347-363 (2019).
- Goessling, W., Sadler, K. C. Zebrafish: An important tool for liver disease research. Gastroenterology. 149 (6), 1361-1377 (2015).
- Huo, X., et al. Transcriptomic profiles of tumor-associated neutrophils reveal prominent roles in enhancing angiogenesis in liver tumorigenesis in zebrafish. Science Reports. 9 (1), 1509 (2019).
- Yan, C., Yang, Q., Gong, Z. Tumor-associated neutrophils and macrophages promote gender disparity in hepatocellular carcinoma in zebrafish. Cancer Research. 77 (6), 1395-1407 (2017).
- Capece, D., et al. The inflammatory microenvironment in hepatocellular carcinoma: A pivotal role for tumor-associated macrophages. Biomed Research International. 2013, 187204 (2013).
- de Oliveira, S., et al. Metformin modulates innate immune-mediated inflammation and early progression of NAFLD-associated hepatocellular carcinoma in zebrafish. Journal of Hepatology. 70 (4), 710-721 (2019).
- Huemer, K., et al. Long-term live imaging device for improved experimental manipulation of zebrafish larvae. Journal of Visualized Experiments. (128), e56340 (2017).
- Huemer, K., et al. zWEDGI: Wounding and entrapment device for imaging live zebrafish larvae. Zebrafish. 14 (1), 42-50 (2017).
- Evason, K. J., et al. Identification of chemical inhibitors of beta-catenin-driven liver tumorigenesis in zebrafish. PLoS Genetics. 11 (7), 1005305 (2015).
- Sapp, V., Gaffney, L., EauClaire, S. F., Matthews, R. P. Fructose leads to hepatic steatosis in zebrafish that is reversed by mechanistic target of rapamycin (mTOR) inhibition. Hepatology. 60 (5), 1581-1592 (2014).
- Park, K. H., Ye, Z. W., Zhang, J., Kim, S. H. Palmitic acid-enriched diet induces hepatic steatosis and injury in adult zebrafish. Zebrafish. 16 (6), 497-504 (2019).
- Progatzky, F., et al. Dietary cholesterol directly induces acute inflammasome-dependent intestinal inflammation. Nature Communication. 5, 5864 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved