Method Article
Analysis of SAMHD1 Restriction by Flow Cytometry in Human Myeloid U937 Cells
In This Article
Summary
Described here is an established method to determine the extent of HIV-1 restriction by the cellular inhibitory protein SAMHD1. Human myeloid lineage U937 cells are transduced with a SAMHD1 expression vector co-expressing YFP, differentiated and then challenged with HIV-RFP. The level of restriction is determined by flow cytometry analysis.
Abstract
Sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) inhibits replication of HIV-1 in quiescent myeloid cells. U937 cells are widely used as a convenient cell system for analyzing SAMHD1 activity due to a low level of SAMHD1 RNA expression, leading to undetectable endogenous protein expression. Based on similar assays developed in the Stoye laboratory to characterize other retroviral restriction factors, the Bishop lab developed a two-color restriction assay to analyze SAMHD1 in U937 cells. Murine Leukaemia Virus-like particles expressing SAMHD1, alongside YFP expressed from an IRES, are used to transduce U937 cells. Cells are then treated with phorbol myristate acetate to induce differentiation to a quiescent phenotype. Following differentiation, cells are infected with HIV-1 virus-like particles expressing a fluorescent reporter. After 48 h, cells are harvested and analyzed by flow cytometry. The proportion of HIV-infected cells in the SAMHD1-expressing population is compared to that in internal control cells lacking SAMHD1. This comparison reveals a restriction ratio. SAMHD1 expression leads to a five-fold reduction in HIV infection, corresponding to a restriction ratio of 0.2. Our recent substitution of RFP for the original GFP as the reporter gene for HIV infection has facilitated flow cytometry analysis.
This assay has been successfully used to characterize the effect of amino acid substitutions on SAMHD1 restriction by transducing with viruses encoding altered SAMHD1 proteins, derived from site-directed mutagenesis of the expression vector. For example, the catalytic site substitutions HD206-7AA show a restriction phenotype of 1, indicating a loss of restriction activity. Equally, the susceptibility of different tester viruses can be determined. The assay can be further adapted to incorporate the effect of differentiation status, metabolic status, and SAMHD1 modifiers to better understand the relationship between SAMHD1, cell metabolic state, and viral restriction.
Introduction
Sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1) impedes replication of Human Immunodeficiency Virus type 1 (HIV-1) in quiescent cells of the myeloid lineage. SAMHD1 blocks replication through its enzymatic activity as a dNTP triphosphohydrolase, which results in decreased levels of intracellular dNTPs. As a result, HIV-1 cannot perform the process of reverse transcription efficiently. Much progress has been made in the few years since this initial observation, particularly regarding the contribution of specific domains and amino acids to SAMHD1's antiviral activity. These insights have been made using biochemical assays as well as cell systems mimicking the physiologically relevant quiescent myeloid environment. U937 cells1 are widely used as a convenient myeloid cell system for analyzing SAMHD1 activity. This is due to a lack of endogenous SAMHD1 expression, thought to be due to low RNA levels compared to SAMHD1-expressing cells (an area of ongoing research). Here, the protocol describes the transient transduction of U937 cells with virus-like particles co-expressing SAMHD1 and a yellow fluorescent protein (YFP) reporter to examine the mechanism of SAMHD1 restriction of HIV-1. Such transient two-color flow cytometry assays to examine retroviral restrictions were first developed in the Stoye laboratory2 and have since been adapted to investigate other restriction factors, including the tripartite motif (TRIM) proteins3. Inspired by these assays, the Bishop lab developed a two-color assay to analyze SAMHD1 restriction in U937 cells.
The workflow for the experiment is shown in Figure 1. Murine Leukaemia Virus (MLV)-like particles containing a bi-cistronic message encoding SAMHD1 alongside YFP expressed from an IRES are used to transduce U937 cells. Cells are then treated with phorbol myristate acetate (PMA) to induce differentiation to a quiescent phenotype. Cells are next infected with HIV-1 virus-like particles expressing red fluorescent protein (RFP). After 48 h, the cells are harvested and analyzed by two-color flow cytometry. This assay is also used with YFP and green fluorescent protein (GFP), requiring less standard filter combinations for flow cytometry analysis. Use of HIV-RFP allows for more straightforward compensation and thus the assay is more easily accessible to less experienced flow cytometry users, and achievable with most cytometers.
During analysis, the proportion of HIV infected cells is compared between SAMHD1-expressing cells and those lacking SAMHD1 within the same well of cells. This affords an internal control in well/flow cytometry tube, which is a key feature. The comparison of infection levels in transduced and untransduced cells reveals a restriction ratio. A ratio of 1.0 indicates that the transduced factor has no effect on infectivity. Wild type SAMHD1 expression leads to a five-fold reduction in HIV infection in this assay, corresponding to a restriction ratio of 0.2. While this effect is modest in comparison to more classic restriction factors such as TRIM5, the effect is nonetheless reproducible and allows the classification of modified SAMHD1 expressors into those that restrict in an equivalent manner to the wild-type protein, those that fail to restrict, and those with an intermediate phenotype.
This assay has been used successfully to characterize SAMHD1 domain and amino acid mutants' restriction phenotypes by transducing with viruses encoding mutant SAMHD1 sequences. For example, the catalytic site substitutions HD206-7AA fail to restrict in this assay. Equally, the susceptibility of different tester viruses can be determined. For example, HIV-1 reverse transcriptase mutants are more susceptible to SAMHD1-mediated restriction (e.g., 151V4). This protocol outlines the details of virus-like particle (VLP) production, transduction of U937 with SAMHD1 expression vectors, infection with HIV carrying a fluorescent reporter and the subsequent analysis by flow cytometry. This article discusses what data to expect and how to avoid suboptimal results. Finally, alternative uses for the assay are outlined, in addition to examining different SAMHD1 variants to understand the interplay between SAMHD1, dNTP levels and viral infection more thoroughly. Given the role of SAMHD1 at the center of cell metabolism, with additional links to cancer, this continues to be an area of intense interest.
Protocol
This protocol does not contain any studies involving animals or human participants performed by any of the authors. All the steps of this protocol were performed following the guidelines and codes of practice of the institutions where they were carried out.
1. Transfection of 293T cells for VLP production (both SAMHD1-expression vectors and tester HIV particles)
NOTE: The most significant contributors to the successful production of virus particles are the health of the producer 293T cells, confluency at transfection and time of harvest. Laboratories will typically have their preferred transfection reagents and protocol for retroviral particle production. The following protocol yields good quality infectious particles, but equivalent protocols would also be suitable for this application. To maintain good quality 293T cells, passage at least three times per week to maintain a constant growth rate. Cells should be seeded in the log phase of growth for efficient transfection.
- Day 1: Seed the required number of 10 cm dishes with a 1/4 split from a near-confluent dish of 293T cells to yield around 60% confluency the following day (approximately 5 x 105 cells/mL). Several densities may need to be seeded to achieve an appropriate density for transfection the following day when working with a new cell stock.
- Day 2 (morning): Check for approximately 60% confluency by microscopy and gently replace media on cells with 10 mL of fresh Dulbecco's Modified Eagle Medium (DMEM).
- Day 2 (afternoon, at least 4 h after media change): Transfect for VLP production.
- Make up the DNA dilution mix containing 3 µg each of gag-pol expressor plasmid, Long-terminal repeat (LTR)-driven reporter plasmid and Vesicular Stomatitis Virus G protein (VSV-G) expressor plasmid (total 9 µg, see NOTE) in 600 µL of serum-free DMEM. Vortex and centrifuge briefly (15 s, >500 x g).
- Add 20 µL of transfection reagent (see Table of Materials), vortex (do not centrifuge) and incubate at 20 °C for 15-20 min. Add the transfection mix dropwise to the plate, swirl gently to mix, and return to the incubator.
NOTE: For MLV VLP expressing SAMHD1 use KB45,6 (gag-pol expressor plasmid), pLGatewaySAMHD1IeYFP1 (LTR-driven reporter plasmid) and pczVSV-G7. For HIV-RFP VLP, use p8.918,9 (gag-pol expressor plasmid), SCRPSY10 (LTR-driven reporter plasmid), and pczVSV-G. Alternatives are discussed in the Table of Materials. Plasmid ratios may need to be optimized for different plasmid/transfection systems.
- Day 3 (late morning): Remove the media carefully from cells by pipetting, wash with 5 mL of fresh DMEM, and replace with 5 mL of fresh DMEM.
- Day 4 (morning): Harvest the virus by collecting the supernatant from the cells and replace with 5 mL of fresh DMEM. Pass the VLP-containing supernatant through a 0.45 nm filter, aliquot and transfer to -70 °C for storage. Ensure that the aliquots are of a size suitable for planned experiments (250 µL as a suggestion) as repeated freeze-thaw cycles will result in a loss of viral titer.
- Day 5 (morning): Harvest the virus as in 1.5 but discard the plates after collecting the supernatant.
2. Titering new VLP prior to use
- Day 1: Seed 293T at 2 x 105 cells per well of a 24-well plate - 4 wells per virus to be titrated, including a virus of known infectivity for each backbone. Optimize the seeding density for a given cell stock.
- Day 2: Observe the plate to check confluency (ideally ~70%). Thaw the virus-containing-supernatant from steps 1.5 and 1.6. Add 0, 10, 25, or 100 µL to each well.
- Day 4 (Morning): Harvest the cells for analysis by flow cytometry.
- Remove the media by careful pipetting or aspiration. Wash the cells gently with 250 µL of phosphate buffered saline (PBS). Remove the cells from the plate by pipetting up and down with 250 µL PBS, and transfer to a microcentrifuge tube. Add 250 µL of 4% paraformaldehyde (taking final concentration to 2%).
CAUTION: Paraformaldehyde is toxic. It should not be mixed with chlorine-based disinfectants. - Pellet the cells by centrifugation at 500 x g for 5 min, discard the supernatant and resuspend the cell pellet in 200 µL of PBS or alternative flow cytometry buffer. Analyze for RFP or YFP as appropriate by flow cytometry. Normalize the infectivity of a given virus stock against values for a stock of known infectivity.
NOTE: VLP can also be normalized by p24 enzyme-linked immunosorbent assay (ELISA); however, this is not necessarily a good indicator of VLP infectivity, especially where VLP stocks are prepared in different transfections. Published median tissue culture infectious dose or multiplicity of infection (TCID50/MOI) methods11 can also provide relative quantitation, although these are not well established for MLV. Relative fluorescence provides a cost-effective alternative to commercial reverse transcriptase ELISA methods.
- Remove the media by careful pipetting or aspiration. Wash the cells gently with 250 µL of phosphate buffered saline (PBS). Remove the cells from the plate by pipetting up and down with 250 µL PBS, and transfer to a microcentrifuge tube. Add 250 µL of 4% paraformaldehyde (taking final concentration to 2%).
3. Transduction of U937 with VLP expressing SAMHD1 and YFP
NOTE: Steps 3-6 constitute the restriction experiment. Days are numbered for ease of planning.
- Day 1 (afternoon): Thaw VLP for transduction including MLV-Wild-type SAMHD1-YFP (positive control), MLV-SAMHD1(HD206-7AA)-YFP (negative control) and variant MLV-SAMHD1-YFP (experimental samples). Dilute the normalized VLP to a final volume of 500 µL with Roswell Park Memorial Institute 1640 media (RPMI) in a microfuge tube, add 0.5 µL of polybrene (10 mg/mL), and mix by flicking.
- If VLP cannot be titrated beforehand, then use 100 µL in the first instance. Use a maximum 1:1 ratio of VLP to RPMI to limit VLP-induced toxicity. Aim for approximately 30% transduction.
- Aliquot 5 x 105 U937 cells into a sterile microfuge tube for each transduction condition. Pellet by centrifugation at 500 x g for 5 min, and remove the supernatant (include two additional tubes as untransduced controls for flow cytometry). Resuspend the cell pellet in 500 µL of diluted VLP (or RPMI for untransduced controls) and transfer to one well of a 24-well plate.
- Spinoculate in a tabletop centrifuge at 800 x g for 90 min at 20 °C. Add 1 mL of 37 °C RPMI to the well and allow to recover in a 37 °C incubator for 3 days.
4. Differentiation of transduced U937
- Day 4 (morning): Re-suspend the cells by gently pipetting and transfer 350 µL into each of 4 wells of a 12-well plate. Add 700 µL of 37 °C RPMI containing 150 nM PMA to each well giving a final concentration of 100 nM and allow to differentiate for 3 days.
NOTE: PMA is bought as a powder and should be resuspended to 1 mM in DMSO and stored in the dark. A working stock of 100 µM should be prepared by diluting 1/10 from the 1 mM stock with DMSO.
5. Infection of differentiated, SAMHD1-expressing U937 with HIV-RFP
- Day 7: Observe the cells by microscopy. Ensure that the cells are adherent. Aspirate the media from all the wells, add 1 mL of RPMI to well 1 of each set of 4 allowing for untransduced, uninfected control as well as single color controls. Add back 0.5 mL of RPMI containing roughly 10 µL of HIV-1-RFP (approximately 10 ng p24 as a guide) to all the other wells. Aim for approximately 50% infection.
- Day 8 (morning): Add 0.5 mL of RPMI to each of the infected wells.
6. Flow cytometry analysis
NOTE: It is good practice to include a live-dead stain in flow cytometry pipelines. However, if the U937 cells are of good quality, this step can be omitted without negative consequences for downstream analysis. Gentle pipetting of trypsinized cells should easily result in a single cell suspension.
- Day 10 (morning): Aspirate the media from the wells and wash once with PBS. Add 300 µL of cell dissociation enzyme (or trypsin) for 5-10 min. Add an additional 300 µL of PBS, resuspend thoroughly making sure all the cells have come off the plate, and transfer to flow cytometry tubes.
- Add 300 µL of 4% paraformaldehyde (taking final concentration to 2%). Pellet the cells by centrifugation at 500 x g for 5 min, discard the supernatant and resuspend the cell pellet in 200 µL of PBS or alternative flow cytometry buffer. Analyze by flow cytometry.
NOTE: The steps below are for a Fortessa analyzer, but equivalent analysis can be achieved using any analyzer capable of reading RFP and YFP fluorescence. Consult local flow cytometry support or other experienced researcher before setting up any flow cytometry analysis. When many samples need to be screened at the same time, a high throughput sampler may be appropriate. In this case, a larger cell number and volume are advised.
7. Flow cytometry analysis
- Turn on the cytometer 10 min before it is required and turn on the computer. Check that the waste is empty and that the sheath tank is full.
- Log in to the machine and open the analysis software.
- Move the arm to the side, take the water tube off, set the flow rate to High and press Prime. Wait for the light to go out and repeat three more times.
- Put the water back on and run on high for 3 min and then switch to Low and press Standby until proceeding to acquisition.
- Meanwhile set up the experiment within the software:
- Select Experiment/ New Experiment (name according to local guidance). In the inspector, delete the unrequired fluorochromes, leaving Blue Laser 530/30 and Yellow Laser 610/20 or the recommended laser and filter combinations for YFP and RFP for the machine.
- Right click on the experiment and choose Cytometer Settings/Application Settings/Create Worksheet. On the worksheet, create a Forward Scatter-Area (FSC-A)/Side Scatter Area (FSC-A) dot blot and YFP histogram, RFP histogram, and GFP/YFP dot blot by clicking on the corresponding icon and changing the axes by clicking on the axis label.
- Right click again on Experiment and add New Specimen. Rename as appropriate. Open the specimen by clicking the plus sign to reveal a new tube.
- Open the acquisition dashboard from View, load the uninfected, untransduced control and press Acquire. Adjust FSC and SSC voltages in the dashboard to position cells in the lower-left quadrant (no cells on the axes). Gate around this population P1. Right click on other plots and select show P1 to remove debris from subsequent analysis.
- Load the single-color control for YFP (wild-type SAMHD1 only) and acquire. Adjust the YFP voltage in the dashboard so that the most fluorescent cells are within the range of the detector (not on the edge of the axis). Repeat for single color RFP control.
- Run automatic compensation using the untransduced, uninfected and single-color controls.
- Select Experiment > Compensation Setup > Create Compensation Controls from the menu to toggle to a normal worksheet. Select unstained normal worksheet, press on Run and acquire for the unstained control.
- Draw a gate around the intact cells (P1) and record. Remove the tube and put the machine on Standby. Right click on P1, click on Apply to All Compensation Controls.
- Switch to the RFP normal worksheet, and press Run. Record the single-color RFP control, and put the machine on Standby. The software should automatically select the positive population.
- Repeat for YFP single color control. Select Experiment > Compensation Setup > Calculate Compensation > Link and Save.
- Switch to the global worksheet. Acquire the cells transduced with wild type SAMHD1, infected with HIV-RFP and check that four distinct populations can be seen in the corners of the quadrants that are aligned vertically and horizontally for YFP and RFP, respectively. Remove the tube and press Standby.
- Reload the untransduced, uninfected sample, press Run and Record 30,000 events with P1 as the stopping gate. Repeat for the remaining samples (including other controls). Rename the samples while running or post hoc. Once exported, files cannot be renamed.
- Export the data as .fcs files and clean the fluidics according to local instructions.
8. Data analysis
NOTE: The steps that follow correspond to analysis in FlowJo v10; however, equivalent steps can easily be performed in other analysis software.
- Open the software and drag all .fcs files into the dashboard. Select the compensation controls and drag them into the compensation subfolder. Open the file for the untransduced, uninfected tube by double-clicking, which will bring up the FSC-A/SSC-A plot.
- Select the polygon tool and gate on the intact cell population, excluding debris in the lower-left corner. Name this Cells. Drag this gate onto the whole population of tubes in the All Samples bar. Scroll through each individual sample using the horizontal arrow buttons to ensure gating is appropriate for each tube.
- Double-click on the Cells population for the untransduced, uninfected sample and adjust the axes to FSC-Height (H) vs FSC-Area (A). Select the single large population using a rectangular gate to exclude doublets. The software will automatically suggest naming this Single Cells.
- Drag this gate onto all the Cells populations and again, check that the gating is appropriate for all samples. Select the Single Cells population for the uninfected, untransduced sample, change the axes to compensated blue laser (y, YFP) versus compensated yellow laser (x, RFP). Press T on the axis and select Biexponential scale for x and y.
- Select the Quadrant Gating Tool and click at the upper-right extreme of the negative cell population. Apply this preliminary gating to all Single Cell populations by dragging into the All Samples bar. Where the quadrant gates do not separate the populations satisfactorily, it may be necessary to gate individual quadrants using the polygon tool, as in Figure 2.
- Scroll to the wild-type SAMHD1 only sample (YFP only) and check that the gating between the untranduced (YFP negative) and YFP positive is correct. It may help to switch to contour view to discriminate negative from dim YFP cells. If the upper-most YFP +ve cells are spread widely, adjust the upper-vertical gate to the right.
- Scroll through all samples and check the gating with respect to YFP positivity. Use the best division between YFP negative and positive that is valid for all samples.
NOTE: The compensation will have been calculated based on the wild-type SAMHD1 YFP expression level. If infection levels are very different between different SAMHD1-YFP transduced cells, the compensation may need to be adjusted. It is therefore preferred that YFP VLP are normalized prior to use. - Scroll to the untransduced, HIV-RFP infected tube and check whether the gating between uninfected RFP negative and infected RFP positive is correct. Adjust as appropriate, using the contour view if required and scroll through the remaining samples to check the gating is valid for all samples.
- Each sample requires double negatives (Q4: lower left, untransduced, uninfected), YFP positive (Q1: upper left, SAMHD1 expressing, uninfected), RFP-positive (Q3: lower right, untransduced, HIV infected), and double positive (Q2: upper right, SAMHD1 expressing HIV-infected). Open the table editor by clicking on the icon and drag the four quadrant gates into the dashboard. Select Excel Export and click on Create Table. Save the generated spreadsheet according to local file naming conventions.
- To export representations of plots and gating strategies, click on the Layout Editor icon. Select each population and drag into the editor with the axes, labeling and spacing required.
- To create a layout with the same plots shown for all samples, select one column and press on Batch. Press on Scale to Width and then Avoid Page Breaks. Save in the file format required.
- In the exported spreadsheet, calculate the restriction ratio by generating columns of percentage RFP of YFP-ves (RFP+ve / (double negatives + RFP+ve)) and percentage RFP of YFP+ves (double positives / (double positives + YFP+ve)) and then a final column of (%RFP of YFP+ve / %RFP of YFP-ve), see Figure 1. Average the replicate data for each SAMHD1 construct and calculate whether restriction values for tested SAMHD1 variants are significantly different from wild-type and/or 206-7AA using appropriate data analysis software.
Results
The results of the analysis above should yield a restriction ratio of 0.25 or lower for the wild-type SAMHD1 positive control, and 1.0 for the negative control. If these two quality control checks are valid, then consider the statistical significance of the results. SAMHD1 variants that show no significant difference from wild type therefore carry substitutions which do not impact SAMHD1 restriction in this context. Those significantly different from wild type show impaired restriction. If these are not significantly different from the negative control, then they lack the ability to restrict in this context (Figure 4 left panels).
If the wild-type SAMHD1 restriction value is greater than 0.3, then results for tester viruses may be indicative but cannot be relied upon. Ineffective restriction by wild-type protein may result from using U937 cells too early post-recovery from reconstitution (within 2 weeks), or when they are too old (>2-3 months). These parameters may need to be empirically determined for a given cell stock. Typically, the lower the passage, the more reliably the cells differentiate and therefore provide the appropriate environment for SAMHD1 restriction. Inappropriate infection level with either SAMHD1-YFP or HIV-RFP may also result in difficulties with both compensation and downstream determination of restriction ratio. An example is illustrated in the right-hand panels of Figure 4.
If the negative control deviates from 1.0 (outside the range of 0.9-1.2), this may indicate a problem with the analysis, either the proportion of infected cells, gating strategy, or health of the cells are affecting the assay. Refer to NOTES above.
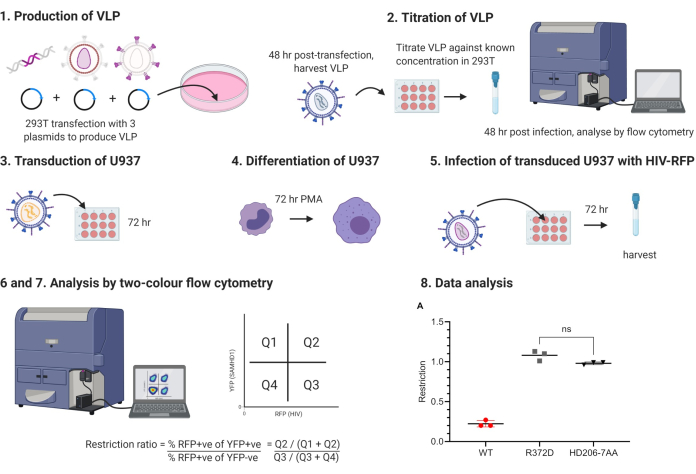
Figure 1: Schematic outlining protocol. VLP, virus-like particles, PMA, phorbol myristate acetate. Numbered stages correspond to stages in protocol. Figure produced using BioRender.com. Please click here to view a larger version of this figure.
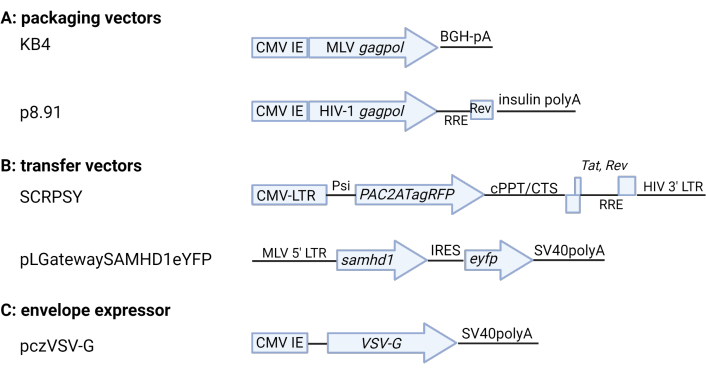
Figure 2: Schematic of retroviral plasmids. (A) Packaging vectors (B) Transfer vectors (C) VSV-G envelope expressor. Key coding and regulatory elements are shown. For details refer to the Table of Materials. CMV IE: cytomegalovirus immediate-early promoter, BGH: bovine growth hormone, pA: polyA, RRE: Rev Response Element, CMV-LTR: composite CMV-HIV-1 LTR promoter, Psi: HIV-1 packaging signal, cPPT/CTS: central polypurine tract/central termination sequence, SV40: simian vacuolating virus 40. Figure produced using BioRender.com. Please click here to view a larger version of this figure.
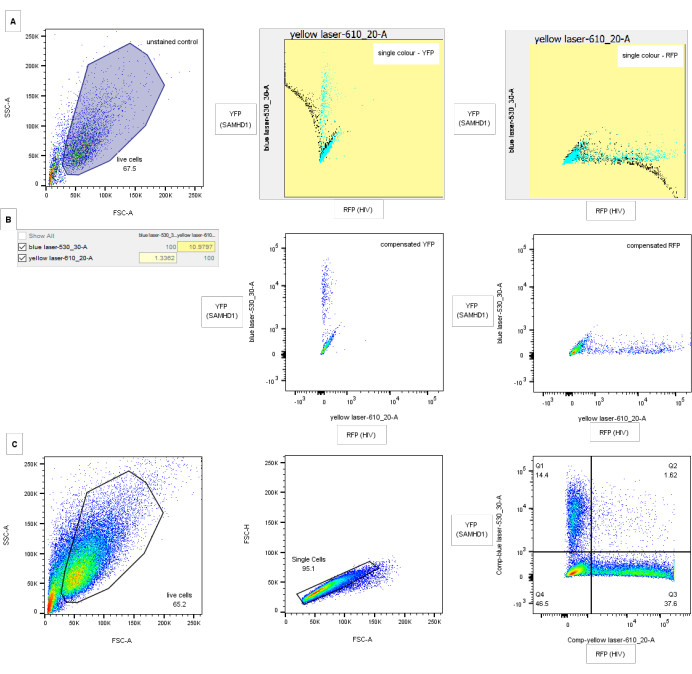
Figure 3: Gating strategy for flow cytometry analysis. (A) Compensation controls: left panel shows FSC-A/SSC-A gating on in-tact cells for the untransduced, uninfected control. Central and right panels show screenshots of single-color compensation controls for YFP (mid) and RFP (right) with uncompensated data in black and compensated in blue. (B) Corresponding compensation matrix and plots for compensation controls above. (C) Gating strategy. Debris is eliminated through analysis of all cells by FSC-A/SSC-A (left panel). Doublets are excluded by gating on FSC height versus area (central panel). Right panel shows an example of HIV-1 restriction by wild-type SAMHD1. Axes show compensated blue and yellow laser fluorescence corresponding to YFP (SAMHD1) and RFP (HIV)-positive cells respectively. Quadrant gates are drawn though comparison of negative and single-color controls for RFP and YFP. Numbers indicate percentage of parent population. Compensation values for YFP with GFP will be much higher but possible to discriminate using appropriate filter sets. Please click here to view a larger version of this figure.
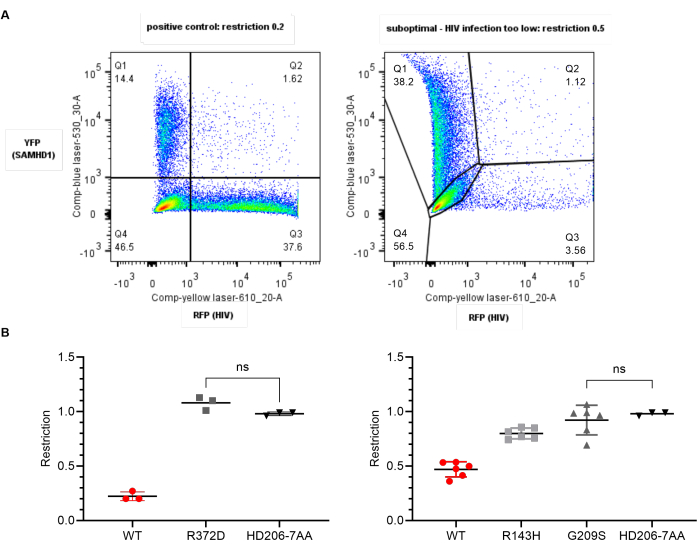
Figure 4: Expected results: ideal versus suboptimal data. (A) Representative YFP/RFP plots for optimal (left) and suboptimal (right) data. Numbers indicate percentage of parent population. In the right panel, HIV infection is too low creating difficulties with compensation and gating. Restriction ratio is higher than expected at 0.5. (B) Plots of restriction ratio for variants of SAMHD1 with respect to wild type (WT, red) or the negative control (HD206-7AA, black) generated using statistical software. Each point represents a replicate value. Mean and standard deviation are shown. Paired t-tests between each group were significant in all cases expect where shown. Left: Ideal data - WT shows the expected restriction of approximately 0.2, negative shows 1.0. The variant R372D (gray) is significantly different from WT but not significant from the negative control and thus has lost the ability to restrict. Right: Suboptimal data. Here, the negative behaves as expected but in all six replicates the WT only shows a restriction ratio of 0.5, due to low infection rate. The low variance within the groups means R143H shows an intermediate phenotype statistically different from WT and the negative, while G209S does not restrict; however, this should be repeated with fresh cells as the positive control has not given the expected restriction ratio. Please click here to view a larger version of this figure.
Discussion
As discussed in the notes above, the critical aspects of the protocol center around retaining the correct cell phenotype for VLP production and for creating the appropriate environment for observing SAMHD1 restriction. Firstly, the accurate analysis of restriction by two-color flow cytometry relies on achieving appropriate proportions of transduced and infected cells so that there are enough cells in each area of the plot for analysis. As such the researcher should be aiming for an MOI of less than 1 (see Protocol). Transduction and infection rates that are either too high or too low lead to problems with analysis due to lack of uninfected or doubly infected cells. Equally, a similar transduction rate for SAMHD1 vectors to be compared is key to allowing the same compensation matrix and gating to be applied to the whole experiment, increasing confidence in the subsequent analysis.
Secondly, the age of the U937 cells used is a key factor in whether they differentiate successfully. Successful differentiation can be monitored through observation of adherence, a reduced acidification of the media over time following differentiation and adequate restriction by the wild type positive control in the assay. Differentiation of up to 5 days has been successful.
One of the key advantages to this assay is its flexibility. A variety of modified versions of SAMHD1 (domains, amino acids, species sequences) can be tested via simple site-directed mutagenesis of the vector or cloning. Equally, variants of the tester virus can be explored. The assay has been used previously to demonstrate that HIV-1 reverse transcriptase mutants with reduced capacity to bind dNTP are more sensitive to SAMHD1 restriction. The same principle can be used to test the restriction of other viruses by SAMHD1. Furthermore, varying differentiation conditions or artificial manipulation of intracellular dNTP concentrations can be used to further probe the interaction between intracellular conditions and SAMHD1 activity, an area of active research in the Bishop lab. The interaction of SAMHD1 with various nucleoside reverse transcriptase inhibitors has also been described, and the effect of SAMHD1 shown using this assay modified for use in primary cells12,13,14,15.
Adaptation of the protocol for use in primary cells overcomes the limitation posed by examining metabolic phenotypes in transformed cells. However, the existing format allows for a convenient and less technically challenging protocol for high throughput analysis, especially with use of a flow cytometer with a high-throughput sampler. When adapting the protocol, the susceptibility of the internally untransduced cells to bystander effects (e.g., immune signaling) should be considered.
As with many aspects of cell biology, it is essential that such assays are used as part of a holistic approach for understanding a given phenomenon. The parallel use of biochemical assays for SAMHD1 activity16, quantification of intracellular dNTP pools, and observation of SAMHD1 mutant phenotypes in humans, ex vivo and in animal models (being carried out by labs around the world, some described in this issue) are key for the evolving understanding of this complex protein.
Disclosures
For the purpose of Open Access, the authors have applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Acknowledgements
This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001042 and FC001162), the UK Medical Research Council (FC001042 and FC001162), and the Wellcome trust (FC001042 and FC001162) and by a Wellcome Trust Investigator Award to JPS (108012/Z/15/Z).Work at the University of Cambridge was co-funded by the NIHR Cambridge Biomedical Research Centre, The Evelyn Trust (16/21), The Rosetrees Trust (M590), and the UK Medical Research Council (MR/S009752/1). The latter UK funded award is part of the EDCTP2 programme supported by the European Union.
Materials
| Name | Company | Catalog Number | Comments |
| 293T cells | ATCC | CRL-3216 | |
| 0.45 µm syringe filters | Sartorius | FCT122 | |
| 10 cm dishes | Corning | 430167 | virus preps can be scaled up through transfection in higher capacity plates/flasks - see transfection reagent guidance |
| 12 well plates | Greiner | 665180 | |
| 24 well plates | Greiner | 662160 | |
| BD LSRFortessa cell analyzer | BD Bioscience | 649225 | alternative analysers can be used as long as they can read RFP and YFP fluorescence |
| DMEM | Sigma | D6429 | ATCC recommends Dulbecco's Modified Eagle's Medium (DMEM) is modified to contain 4 mM L-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, and 1500 mg/L sodium bicarbonate. |
| DMSO | Fisher | BP-231-100 | |
| fetal bovine serum | Gibco | 10500064 | |
| Flowjo | BD Bioscience | - | alternative analysis software can be used |
| light microscope | - | - | Any bright field light microscope |
| p8.91 | - | - | HIV GagPol expressor (PMID: 8602510) |
| paraformaldehyde (4 % in PBS) | Alfa Aesar | J61889 | |
| PBS | Sigma | D8537 | |
| Penicillin-Streptomycin (10,000 U/mL) | ThermoFisher | 15140122 | |
| phorbol 12-myristate 13-acetate (PMA) | Sigma | P8139 | |
| polybrene | Sigma | TR-1003 | |
| pKB4 | - | - | Murine leukaemia Virus GagPol expressor (PMID: 23035841) |
| pLGatewaySAMHD1eYFP | - | - | MLV packaging plasmid encoding wild-type human sequence. Details and cloning procedures Arnold et al 2015 (Ref 1). |
| pLGatewaySAMHD1HD206-7AAeY FP | - | - | As above, encodes amino acid substitutions at crucial active sites residues |
| Prism | Graphpad | - | https://www.graphpad.com/scientific-software/prism/ Other data representation software are also suitable |
| pVSV-G | - | - | VSV-G expressor (https://www.addgene.org/138479/), alternative: pMD2.G (https://www.addgene.org/12259/) |
| RPMI | ThermoFisher | 21875034 | |
| screw-cap tubes | StarLab | E1415-2231 | |
| SCRPSY | - | - | HIV packaging plasmid encoding RFP, Sam Wilson, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5026698/ |
| stripettes 10 mL | Corning | 10084450 | |
| stripettes 5 mL | Corning | 10420201 | |
| TrypLE express | Gibco | 12604021 | Trypsin will also be suitable |
| Turbofect | ThermoFisher | R0531 | alternative transfection reagents will also work well |
| U937 cells | ATCC | CRL-1593.2 |
References
- Sundström, C., Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). International Journal of Cancer. 17 (5), 565-577 (1976).
- Bock, M., Bishop, K. N., Towers, G., Stoye, J. P. Use of a transient assay for studying the genetic determinants of Fv1 restriction. Journal of Virology. 74 (16), 7422-7430 (2000).
- Yap, M. W., Nisole, S., Lynch, C., Stoye, J. P. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proceedings of the National Academy of Sciences of the United States of America. 101 (29), 10786-10791 (2004).
- Arnold, L. H., et al. Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathogens. 11 (10), 1005194 (2015).
- Groom, H. C. T., et al. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology. 7, 10 (2010).
- Mothes, W., Boerger, A. L., Narayan, S., Cunningham, J. M., Young, J. A. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 103 (4), 679-689 (2000).
- Pietschmann, T., et al. Foamy virus capsids require the cognate envelope protein for particle export. Journal of Virology. 73 (4), 2613-2621 (1999).
- Naldini, L., et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 272 (5259), 263-267 (1996).
- Bainbridge, J. W., et al. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Therapy. 8 (21), 1665-1668 (2001).
- Kane, M., et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host and Microbe. 20 (3), 392-405 (2016).
- Gao, Y., et al. Calculating HIV-1 infectious titre using a virtual TCID50 method. Methods in Molecular Biology. 485, 27-35 (2009).
- Ordonez, P., et al. SAMHD1 enhances nucleoside-analogue efficacy against HIV-1 in myeloid cells. Scientific Reports. 7, 42824 (2017).
- Ballana, E., et al. SAMHD1 specifically affects the antiviral potency of thymidine analog HIV reverse transcriptase inhibitors. Antimicrobial Agents and Chemotherapy. 58 (8), 4804-4813 (2014).
- Huber, A. D., et al. SAMHD1 has differential impact on the efficacies of HIV nucleoside reverse transcriptase inhibitors. Antimicrobial Agents and Chemotherapy. 58 (8), 4915-4919 (2014).
- Amie, S. M., et al. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. The Journal of Biological Chemistry. 288 (28), 20683-20691 (2013).
- Arnold, L. H., Kunzelmann, S., Webb, M. R., Taylor, I. A. A continuous enzyme-coupled assay for triphosphohydrolase activity of HIV-1 restriction factor SAMHD1. Antimicrobial Agents and Chemotherapy. 59 (1), 186-192 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved