A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
High-Pressure NMR Experiments for Detecting Protein Low-Lying Conformational States
In This Article
Summary
We provide a detailed description of the steps required to assemble a high-pressure cell, set up and record high-pressure NMR experiments, and finally analyze both peak intensity and chemical shift changes under pressure. These experiments can provide valuable insights into the folding pathways and structural stability of proteins.
Abstract
High-pressure is a well-known perturbation method that can be used to destabilize globular proteins and dissociate protein complexes in a reversible manner. Hydrostatic pressure drives thermodynamical equilibria toward the state(s) with the lower molar volume. Increasing pressure offers, therefore, the opportunities to finely tune the stability of globular proteins and the oligomerization equilibria of protein complexes. High-pressure NMR experiments allow a detailed characterization of the factors governing the stability of globular proteins, their folding mechanisms, and oligomerization mechanisms by combining the fine stability tuning ability of pressure perturbation and the site resolution offered by solution NMR spectroscopy. Here we present a protocol to probe the local folding stability of a protein via a set of 2D 1H-15N experiments recorded from 1 bar to 2.5 kbar. The steps required for the acquisition and analysis of such experiments are illustrated with data acquired on the RRM2 domain of hnRNPA1.
Introduction
It has long been recognized that higher-energy, sparsely populated conformational states of proteins and protein complexes play a key role in many biological pathways1,2,3. Thanks to experiments based on Carr-Purcell-Meiboom-Gill (CPMG)4, Chemical Exchange Saturation Transfer (CEST)5, and dark-state exchange saturation transfer (DEST)6 pulse sequences (among others), solution NMR spectroscopy has emerged as a method of choice for characterizing transient conformational states7. Along with these experiments, perturbations such as temperature, pH, or chemical denaturants can be introduced to increase the relative population of higher energy conformational substates. Similarly, protein equilibria can also be perturbed by applying high hydrostatic pressure. Depending on the magnitude of the volume change associated with the corresponding conformational changes, an increase of pressure by a few hundred to a few thousand bars can significantly stabilize a higher energy state or cause a protein to completely unfold8,9,10. Protein NMR spectra typically display two types of changes with hydrostatic pressure: (i) chemical shift changes and (ii) peak intensity changes. Chemical shift changes reflect changes at the protein surface-water interface and/or local compression of the protein structure on a fast time scale (relative to NMR time scale)11. Crosspeaks exhibiting large non-linear chemical shifts pressure dependence can indicate the presence of higher energy conformational states12,13. On the other hand, peak intensity changes point to major conformational transitions on a slow time scale, such as changes in folded/unfolded state populations. The presence of folding intermediates or higher energy states can be detected from large variations in the magnitude of the volume change upon unfolding measured for different residues of a given protein14,15,16,17. Based on our experience, even small proteins that are typically classified as two-state folders exhibit non-uniform responses to pressure, which provides useful information about their local folding stability. Described here is a protocol for the acquisition and analysis of amide peak intensity and 1H chemical shifts pressure dependence, using as a model protein the isolated RNA recognition motif 2 (RRM2) of the heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1).
Access restricted. Please log in or start a trial to view this content.
Protocol
NOTE: The protocol described here requires (i) a high-pressure pump and cell with a 2.5 kbar rated aluminum-toughened zirconia tube18, (ii) the software SPARKY19 for analysis of the NMR spectra, and (iii) a curve fitting software.
1. Sample preparation, assembly of the high-pressure cell, and setting up the experiments.
- Choice of buffer: Use equal mixture of anionic and cationic buffers, such as phosphate and Tris20,21.
NOTE: The pKa of anionic buffers such as phosphate and MES is associated with a substantial reaction volume (i.e., the difference in the partial molal volumes of the acid and the ionized products). The pH of such buffers can therefore be significantly affected by a change of pressure (~0.25-0.5 pH unit/kbar). - Ensure that the required sample volume is similar to that of a standard 3 mm diameter NMR tube (~300μL).
- Introduce the 15N-labeled sample with a glass pipette into the zirconia tube. Make sure the sample seats at the bottom of the tube. Complete with 200 μL of mineral oil to prevent the sample from mixing with the transmission liquid (e.g., water). Fill the rest of the tube with transmission liquid.
- Put a single-use O-ring on top of the zirconia tube and slide the tube into the base (Figure 1A,B). Then, connect the tube to the high-pressure tether line and tighten the base to the cell first by hand. Then, apply 14.7 Nm of torque to prevent leaks at lower pressure (Figure 1C,D).
- To check the integrity of the pressure cell assembly, pressurize the tube up to 300 bar outside the spectrometer using cell support and containment vessel. Wait for 15 min, reset the pressure to 1 bar and check for leaks with a clean lint-free wipe.
- Insert the unpressurized tube into the spectrometer by carefully guiding the tether line. Slide the tube in the spectrometer until reaching the sample sitting position (Figure 1E).
- Lock, shim, match, and tune the 1H and 15N channels as usual.
NOTE: Shims for high-pressure rated zirconia tubes are very different from standard NMR tubes. It is recommended to save the optimized shims for future use. - Set up a 1H-15N-HSQC or TROSY-HSQC and record a reference experiment at atmospheric conditions (1bar).
2. Recording high-pressure NMR experiments
- Gradually increase the pressure from 1 bar to 2.5 kbar with 500 bar increments to test the overall stability of the protein. Set the speed of the pressure pump by default at ~18 bar/s. If the precise folding/unfolding rates are not known, let the sample equilibrate 15-20 min after each 500-bar increment. Record a spectrum at 2.5 kbar.
- Gradually decrease the pressure back to 1 bar with 500 bar steps to test the reversibility of the pressure perturbation. Record another spectrum at atmospheric conditions and compare the chemical shifts and peak intensities with that of the reference spectrum previously recorded in the same conditions.
NOTE: If the native crosspeaks are more intense after the pressure run, it may indicate that small aggregates present in solution at atmospheric pressure may have dissociated and properly refolded. On the other hand, a loss in the intensity or significant chemical shift changes suggest that the protein may experience a non-reversible misfolding in high-pressure conditions. - Record a series of 2D experiments from 1 bar to 2.5 kbar every 500 bar. It is recommended to record additional experiments near the inflection point of the folding/unfolding transition to improve the precision of the fit.
3. Analyzing peak intensity changes
- Process all the spectra and transfer the backbone assignment from the reference spectrum at 1 bar to the spectrum recorded at 500 bar, and then transfer the assignment from 500 bar to 1 kbar and so forth.
NOTE: Because pressure induces a non-uniform shift of 1H and 15N chemical shifts, do not simply copy the backbone assignment from one spectrum to the next. Adjust it manually. - In Sparky menu, click on Peak > Peak List (lt). In the Peak List window, click on Options and select the option to display both Frequencies (ppm) and Data Height. Save the list obtained for each spectrum.
- In a curve fitting software, copy the crosspeak identity and peak intensity values to have the pressure values (in bar) as the X-axis variable and the intensity as the Y-axis variable.
- If a complete or near complete (>80%) unfolding is observed, fit the individual peak intensity profiles to extract the free energy and volume change upon unfolding, respectively, using a simple two-state model:
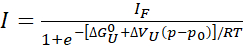 Eq. 1
Eq. 1
Where, "I" is the observed intensity of a crosspeak at a given pressure p and IF is the intensity of the same crosspeak in a fully folded state. R is the gas constant, T is the absolute temperature, ΔGU0 the standard Gibbs free energy difference between the unfolded and folded states at atmospheric pressure p0 (1 bar), and ΔVu the volume change upon unfolding. With the pressure p in bar and temperature T in kelvin, R = 1.987 cal/K, ΔGU0 is in cal/mol, and ΔVU is in cal/mol/bar. Multiply the ΔVU values obtained from the fit by 41.84 to convert into mL/mol. ΔVU values are typically in the range of -50 to -150 mL/mol for globular proteins22. Here, all the parameters are expressed with regard to the unfolding reaction but can easily be converted into folding reaction parameters (ΔGU0 = -ΔGF0 and ΔVU = -ΔVF).
4. Analyzing chemical shift changes
- Arrange the columns in the software in order to have pressure points as variable and the 1H chemical shifts extracted from Sparky lists as the Y-axis.
- Fit the pressure dependence of the 1H chemical shifts to a simple quadratic equation:
δ(p) = δ0(p0) + B1(p-p0) + B2 (p-p0)2 Eq. 2
Where, δ(p) is the measured 1H chemical shift of a crosspeak at a given pressure p and δ0(p0) the 1H chemical shifts of the same crosspeak in the reference spectrum recorded at 1 bar. B1 and B2 represent the first and second-order parameters expressed in ppm/bar and ppm/bar2, respectively.
Access restricted. Please log in or start a trial to view this content.
Results
The protocol described here was used to probe the pressure dependence of RRM2, the second RNA recognition motif of hnRNPA1 (residues 95-106), which is almost completely unfolded within the 2.5 kbar range (>90%). 1H-15N spectra were collected at 1 bar, 500 bar, 750 bar, 1 kbar, 1.5 kbar, 2 kbar, and 2.5 kbar (Figure 2). Since none of the native crosspeaks were visible above the noise level at 2.5 kbar, all corresponding residues were attributed an intensity valu...
Access restricted. Please log in or start a trial to view this content.
Discussion
This study details a protocol implemented to probe protein structural and thermodynamics responses to pressure perturbation. The high-pressure experiments recorded here on RRM2 demonstrate that large variations in ΔVU values, indicative of non-fully cooperative unfolding, can be found in a relatively small single domain protein. A similar picture emerges from the analysis of 1H chemical shift changes under pressure. It should be noted Kalbitzer and coworkers have demonstrated that a more i...
Access restricted. Please log in or start a trial to view this content.
Disclosures
All the authors have read and approved the manuscript. They declare no conflicts of interest.
Acknowledgements
This work was supported by funds from the Roy J. Carver Charitable Trust to Julien Roche. We thank J. D. Levengood and B. S. Tolbert for kindly providing the RRM2 sample.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Bruker Nmr Cell 2.5 Kbar | Daedalus Innovations LLC | NMRCELL-B | |
| Sparky3 | University of California San Francisco, CA | N/A | |
| Xtreme-60 Syringe pump | Daedalus Innovations LLC | XTREME-60 |
References
- Alderson, R. T., Kay, L. E. Unveiling invisible protein states with NMR spectroscopy. Current Opinion in Structural Biology. 60, 39-49 (2020).
- Korzhnev, D. M., Kay, L. E. probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: An application to protein folding. Accounts of Chemical Research. 41, 442-451 (2008).
- Loria, P. J., Berlow, R. B., Watt, E. D. Characterization of enzyme motions by solution NMR relaxation dispersion. Accounts of Chemical Research. 41, 214-221 (2008).
- Ishima, R. CPMG relaxation dispersion. Methods in Molecular Biology. 1084, 29-49 (2014).
- Longo, D. L., et al. Chemical exchange saturation transfer (CEST): an efficient tool for detecting molecular information on proteins' behaviour. Analyst. 39, 2687-2690 (2014).
- Fawzi, N. L., Ying, J., Torchia, D. A., Clore, M. G. Probing exchange kinetics and atomic resolution dynamics in high-molecular-weight complexes using dark-state exchange saturation transfer NMR spectroscopy. Nature Protocols. 7, 1523-1533 (2012).
- Anthis, N. J., Clore, M. G. Visualizing transient dark states by NMR spectroscopy. Quarterly Reviews of Biophysics. 48, 35-116 (2015).
- Roche, J., et al. Cavities determine the pressure unfolding of proteins. Proceedings of the National Academy of Sciences of the United States of America. 109, 6945-6950 (2012).
- Chen, C. R., Makhatadze, G. I. Molecular determinant of the effects of hydrostatic pressure on protein folding stability. Nature Communications. 8, 14561(2017).
- Roche, J., Royer, C. A. Lessons from pressure denaturation of proteins. Journal of the Royal Society Interface. 15, 20180244(2018).
- Xu, X., Gagné, D., Aramini, J. M., Gardner, K. H. Volume and compressibility differences between protein conformations revealed by high-pressure NMR. Biophysical Journal. 120, 924-935 (2021).
- Akasaka, K., Li, H. Low-lying excited states of proteins revealed from non-linear pressure shifts in 1H and 15N NMR. Biochemistry. 40, 8665-8671 (2001).
- Akasaka, K. Probing conformational fluctuation of proteins by pressure perturbation. Chemical Reviews. 106, 1814-1835 (2006).
- Kitahara, R., Yokoyama, S., Akasaka, K. NMR snapshots of a fluctuating protein structure: ubiquitin at 30 bar-3 kbar. Journal of Molecular Biology. 347, 277-285 (2005).
- Roche, J., et al. remodeling of the folding free energy landscape of Staphylococcal nuclease by cavity-creating mutations. Biochemistry. 51, 9535-9546 (2012).
- Nucci, N. V., Fuglestad, B., Athanasoula, E. A., Wand, J. A. Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proceedings of the National Academy of Sciences of the United States of America. 111, 13846-13851 (2014).
- Maeno, A., et al. Cavity as a source of conformational fluctuation and high-energy state: High-pressure NMR study of a cavity-enlarged mutant of T4 lysozyme. Biophysical Journal. 108, 133-145 (2015).
- Peterson, R. W., Wand, J. A. Self-contained high-pressure cell, apparatus, and procedure for the preparation of encapsulated proteins dissolved in low viscosity fluids for nuclear magnetic resonance spectroscopy. Review of Scientific Instruments. 76, 094101(2005).
- Goddard, T. D., Kneller, D. G. Sparky 3. , University of California San Francisco. San Francisco, CA. (2010).
- Caro, J. A., Wand, J. A. Practical aspects of high-pressure NMR spectroscopy and its applications in protein biophysics and structural biology. Methods. 148, 67-80 (2018).
- Kitamura, T., Itoh, J. Reaction volume of protonic ionization for buffering agents. Prediction of pressure dependence of pH and pOH. Journal of Solution Chemistry. 16, 715-725 (1987).
- Royer, C. A. Revisiting volume changes in pressure-induced protein unfolding. Biochimica et Biophysica Acta. 1595, 201-209 (2002).
- Erlach, M. B., et al. Relationship between nonliner pressure-induced chemical shift changes and thermodynamic parameters. Journal of Physical Chemistry B. 118, 5681-5690 (2014).
- de Oliveira, G. A. P., Silva, J. L. A hypothesis to reconcile the physical and chemical unfolding of proteins. Proceedings of the National Academy of Sciences of The United States of America. 112, 2775-2784 (2015).
- Nguyen, L. M., Roche, J. High-pressure NMR techniques for the study of protein dynamics, folding and aggregation. Journal of Magnetic Resonance. 277, 179-185 (2017).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved