Method Article
Enhancing Tumor Content through Tumor Macrodissection
In This Article
Summary
This protocol presents a method to increase the percent tumor content of formalin-fixed paraffin-embedded tissue samples.
Abstract
The presence of contaminating non-tumor tissues in formalin-fixed paraffin-embedded (FFPE) tissues can greatly undermine genomic studies. Herein we describe macrodissection, a method designed to augment the percentage tumor content of a tissue specimen by removing and eliminating unwanted tissue prior to performing downstream nucleic acid extractions. FFPE tissue blocks were sectioned to produce 4-5 µm slide-mounted tissue sections. A representative section was submitted for hematoxylin and eosin (H&E) staining and subsequently reviewed by a board-certified pathologist. During the review, the pathologist identified and marked the regions of tumor tissue in the H&E. Once complete, the demarked H&E was used to guide resection of the serial unstained sections from the same tissue block. To demonstrate the effects of macrodissection, RNA extracted from matched macrodissected and non-dissected Diffuse Large B-Cell Lymphomas (DLBCL) were run on a digital gene expression assay capable of determining DLBCL subtype and BCL2 translocation status. The results showed that macrodissection changed the subtype or BCL2 translocation status calls in 60% of the samples examined. In conclusion, macrodissection is a simple and effective method for performing tumor enrichment prior to nucleic acid extractions, the product of which can then be confidently used in downstream genomic studies.
Introduction
Formalin-fixed paraffin-embedded (FFPE) tissues, collected as part of the normal clinical diagnostic process and retained in clinical tissue repositories, represent a vast resource for human research, including cancer research1. As our understanding of human disease deepens, it is becoming increasingly clear that diseases, previously thought to be single entities based on morphological and immunophenotypical characteristics, are in fact comprised of distinct molecular subtypes that require molecular subtyping assays. Consequently, high sensitivity genomic assays capable of discerning these subtypes have become increasingly important2. Although FFPE tissues are renowned for being poorly compatible with genomic techniques due to fixation-related issues, as technology and protocols evolve, these techniques are becoming increasingly compatible with this clinically ubiquitous tissue format3,4,5. However, FFPE tissues are often admixtures of tumor and non-tumor tissue materials, where the presence of non-tumor material is frequently unwanted and can, if present at a high proportion, significantly undermine and impact the results of genomic analyses6. Indeed, a minimum tumor content of 60% is frequently used for such analyses, where tissues that fall short of this threshold can be excluded, despite otherwise fulfilling the study criteria7. This can be particularly problematic in rare disease settings, where patient tissues are precious and difficult to collect in high numbers.
Macrodissection is a method that minimizes the effects of low tumor content by reducing the amount of normal tissue3. The removal of such confounding non-tumor material prior to nucleic acid extraction can significantly augment the tumor percentage content and thus the tumor purity of the extracted nucleic acids. Tissue resection critically relies upon expert pathological review, wherein the tumor region is identified and circled on a freshly generated hematoxylin and eosin (H&E) stained tissue section by a board-certified pathologist8. The circled H&E is then used to guide the removal and collection of unwanted and target tissues, respectively. This protocol describes the steps of macrodissection from pathological review through tissue harvesting as performed at the AIDS and Cancer Specimen Resource (ACSR) Technical Core Laboratory at the Mayo Clinic.
Protocol
All samples were collected and used in compliance with approved Mayo Clinic IRB protocols (PR16-000507 and PR2207-02).
1. Sample preparation
- Turn on the tissue floatation water bath. Set the temperature to 39 °C and allow the water to come to temperature. Soak wooden collection sticks in the water bath.
- Identify and retrieve the FFPE tissue blocks to be sectioned.
- Pre-label microscope slides using a histology grade permanent marker that can withstand solvent washes.
- Use a microtome to section the FFPE blocks. Cut at least 2 full-face sections at a thickness of 4-5 μm per section for each block (Figure 1A).
- Transfer the freshly cut ribbon of tissue sections to the pre-warmed tissue floatation bath for slide mounting (Figure 1B, C).
NOTE: The warm water helps to "iron" out the wrinkles in the sections (Figure 1C). - Handling each section sequentially, use forceps to break a single section away from the ribbon (Figure 1D).
- Collect the single section on a pre-labeled microscope slide.
- Submerge the microscope slide at an angle underneath the section, positioning the slide such that the edge of the tissue section touches the slide (Figure 1E).
- Once in contact with the tissue section, pull the slide out of the floatation bath slowly to allow the tissue section to straighten out flush against the slide as it emerges from the water (Figure 1F).
- Mount 1 tissue section per slide and repeat until all the sections have been collected.
- Allow the slide-mounted tissue sections to dry thoroughly at room temperature (RT).
2. Pathological review
- Perform H&E staining on one representative tissue section for each block9.
- Submit the freshly stained H&Es for pathological review by a board-certified pathologist.
NOTE: During the review, the pathologist determines and records the percentage tumor content in each tissue and circles the tumor area on each H&E slide (see Figure 2 and Figure 3, Row 1). Table 1 outlines the percentage tumor content by cellularity determined during the pathological review of the H&Es for samples A-E shown in Figure 3, Row 1. Sections with <60% tumor content require macro-dissection7. The number of sections needed for nucleic acid extraction depends on the size of the circled tumor area. If insufficient sections were cut in section 1 of the protocol and further cuts are possible, then additional sections may need to be cut.
3. Deparaffinization
- In a fume hood, prefill two glass staining dishes with undiluted histology grade d-Limonene or a d-Limonene based solvent and 1 glass staining dish with undiluted 200-proof molecular grade ethanol.
CAUTION: Avoid d-Limonene contact with skin and eyes, avoid inhalation of vapor or mist, and keep away from sources of ignition. Keep ethanol away from heat, sparks, and open flames, avoid spilling and contact with the skin or eyes, ventilate well and avoid breathing vapors.
NOTE: Fill the dishes sufficiently to submerge the racked slides (Figure 4A); 250 mL is required to fill the 20-slide staining dishes shown. Replace the d-Limonene and ethanol washes after every 40 slides. D-Limonene (C10H16) is an alternative, similarly effective, and less toxic dewaxing agent to xylene that is becoming more commonplace in histological methodologies and yields good quality nucleic acid post extraction10,11,12,13,14. Although this protocol may be performed using more biofriendly alternatives15, their effects, if any, on extracted nucleic acid quality remains to be determined. - Rack the unstained FFPE tissue mounted slides into the glass Coplin slide-racks (Figure 4A, inset).
- Submerge the racked slides in d-Limonene wash 1 for 2 min; gently agitate for the first 20 s.
NOTE: To minimize carryover between washes, when removing the rack of slides from a wash, allow the rack to drain briefly before gently dabbing the bottom of the rack on tissue paper to remove excess wash. - Submerge the racked slides in undiluted D-Limonene wash 2 for 2 min; gently agitate for the first 20 s. Remove, drain and dab the rack again.
- Submerge the racked slides in the ethanol wash for 2 min; gently agitate for the first 20 s. Remove the rack and place it on an absorbent tissue to drain. Allow the slides to air dry for at least 10 min, but no more than 2 h.
3. Macrodissection
- On the bench, prefill a Coplin glass jar with 50 mL of 3% glycerol in DNase/RNase free water
NOTE: Replace the glycerol wash after every 40 slides. - Pre-label and prefill 1.5 mL microtubes with 160 µL of tissue digestion buffer per microtube.
NOTE: Nucleic acid extractions performed after macrodissection used a DNA/RNA FFPE extraction kit (Table of Materials). Thus, the tissue digestion buffer used in this protocol comprised 10 µL of Proteinase K and 150 µL of PKD buffer. - Trace the pathological markings on the H&E onto the back of the deparaffinized tissue slides.
NOTE: Compared to non-deparaffinized tissues (Figure 3, Row 2 and Figure 4B), deparaffinized tissues are white and highly visible (Figure 3, Row 3, and Figure 4C). It is this heightened visibility and discernability of deparaffinized tissue features that permit macrodissection. Place the H&E face down on the bench and place the front of the deparaffinized slide against the back of the matched pathologist reviewed H&E (Figure 4D). Align the deparaffinized tissue with the H&E tissue (Figure 4E). Tracing the pathologist's markings is a critical step in the macrodissection process, and care should be taken to reproduce these markings as accurately as possible. This can be particularly challenging for small and or disconnected tissues such as samples B, D, and E (Figure 3, Figure 4E, and Figure 4E inset i). To aid tracing, one should use an inky fine or ultrafine nibbed marker (Figure 4E, inset ii) to trace the pathologist-drawn markings. Ethanol wipes may be useful to remove errors and permit retracing if needed. - Turn the now marked deparaffinized slide tissue face-up and trace the line of the marker with the corner of a clean razor blade to pre-cut the edges of the tumor area.
- Handling each slide sequentially, dip the deparaffinized slides into the 3% glycerol solution. Ensure the tissue is completely submerged before removing the slide slowly (Figure 4F).
NOTE: The purpose of the glycerol dip is to dampen the tissue to aid tissue collection but also to reduce the build-up of static charge that can cause repulsion between the tissue and the plastic microtube in which the collected tissue must be placed. - Gently wipe the back of the slide with a tissue to remove the excess glycerol solution, and lay the slide on the bench, wiped side face down. Allow tissues to briefly airdry for 1-2 min.
NOTE: Carry over of excess glycerol into the extraction process can negatively affect the yield and quality of nucleic acid extractions. Tissues should be slightly damp but not visibly wet when collected. - Depending on where the tumor area of interest is situated on the slide, use the flat edge of the razor blade to either (a) directly collect the tumor tissue, using the razor to scrape/collect the tissue of interest off the slide, or (b) remove and discard non-tumor tissue first before collecting the tumor tissue of interest (Figure 3).
NOTE: Collected tissue tends to gather or roll up at the bottom of the blade (Figure 4G) - Use a wooden stick to remove the collected tissue from the blade (Figure 4H and inset) and transfer it to the appropriate pre-labeled and prefilled microtube (Figure 4I).
NOTE: The digestion buffer serves to "pull" the tissue from the wooden pick into the liquid. - Proceed to nucleic acid extraction.
NOTE: Nucleic acid extractions were completed using the DNA/RNA FFPE extraction kit per the manufacturer's instructions, and the resulting nucleic acids were quantified using a UV-vis spectrophotometer. The resulting RNA was run on the digital gene expression profiling-based DLBCL90 assay16.
Results
A total of 5 Diffuse Large B-Cell Lymphoma (DLBCL) FFPE tissue blocks were sectioned, and the resulting sections were either macrodissected or not prior to nucleic acid extraction. The extracted RNA was run on the DLBCL90 assay16. Macrodissected samples were run twice, once using 5 µL of RNA stock concentration but no more than 300 ng of total RNA input and once using 5 µL of RNA stock diluted to match the RNA inputs of their respective non-dissected counterparts. The DLBCL90 results are outlined in Table 2.
DLBCL is comprised of 3 distinct cell of origin (COO) subtypes with different therapeutic responsiveness, namely GCB, ABC, and an intermediate group known as unclassified or UNC17,18. Translocations involving MYC, BCL2, and or BCL6 alone or in combination (double or triple hit) are also frequently observed in DLBCL, particularly in the GCB subtype19. The DLBCL90 assay is an expansion of its predecessor, the Lymph2Cx clinical assay, and is thus capable of DLBCL COO subtype determination but was principally developed to identify samples harboring double hit (DH) translocations involving BCL2 using digital gene expression as an alternative to fluorescence in-situ hybridization (FISH)16,20. The results in Table 2 show that macrodissection changed either the COO or the DHITsig status calls for 60% (3/5) of the samples examined.
Macrodissection of sample A had no effect on the COO call but changed the DHITsig call from NEG to UNCLASS, and this change was observed irrespective of the macrodissected sample RNA input and with similar probability scores (0.224, 0.254). In contrast, macrodissection of sample C had no effect on the DHITsig call but changed the COO call from GCB to UNC. Again, this change was observed irrespective of macrodissected sample RNA input. However, at 0.117, the COO call probability was closer to the call threshold of 0.1 for the macrodissected sample with reduced RNA input. Similar to sample A, macrodissection of sample E had no effect on the COO call but changed the DHITsig call. However, for sample E the call changed from UNCLASS to NEG and did so irrespective of the macrodissected sample RNA input with reasonably similar probability calls (0.849, 0.833). Notably, this call change to DHITsig NEG makes biological sense given that sample E was found to ABC-DLBCL, and double hit translocations involving BCL2 have been reported to be exclusively observed in GCB-DLBCL19.

Figure 1: Generating slide-mounted tissue sections using a microtome. (A) FFPE tissue block held in place by the microtome chuck and cut to produce a ribbon of sequential FFPE tissue sections. (B) Using pre-soaked wooden sticks, the ribbon is collected from the microtome and transferred to a warm water bath. (C) The warmth of the water helps to iron out the creases in the tissue ribbon. (D) Individual FFPE tissue sections are removed from the tissue ribbon by placing closed forceps at the junction of two sections and gently opening the forceps, which breaks sections away from one another. (E) Sections are collected from the water by submerging a glass slide at an angle and gently moving the side towards the tissue section until the edge of the section touches the glass slide. (F) Once the slide and section are touching, slowly remove the slide from the water, allowing the tissue section to fall flush against the slide. Please click here to view a larger version of this figure.
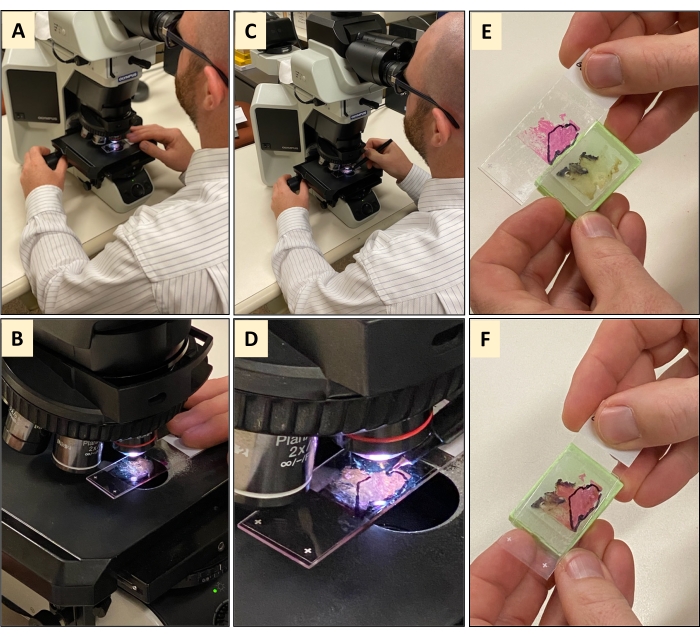
Figure 2: Pathology and histological review of a hematoxylin and eosin (H&E) stained sections. (A,B) The H&E tissue section is subjected to microscopic review by a board-certified pathologist. (C,D) Once the pathologist has viewed the entire tissue and found that it is not 100% tumor tissue, the pathologist will use a marker to encircle the tumor area of the tissue. (E,F) Holding the marked H&E against the original FFPE tissue block shows that not all the tissue is tumor material. Please click here to view a larger version of this figure.
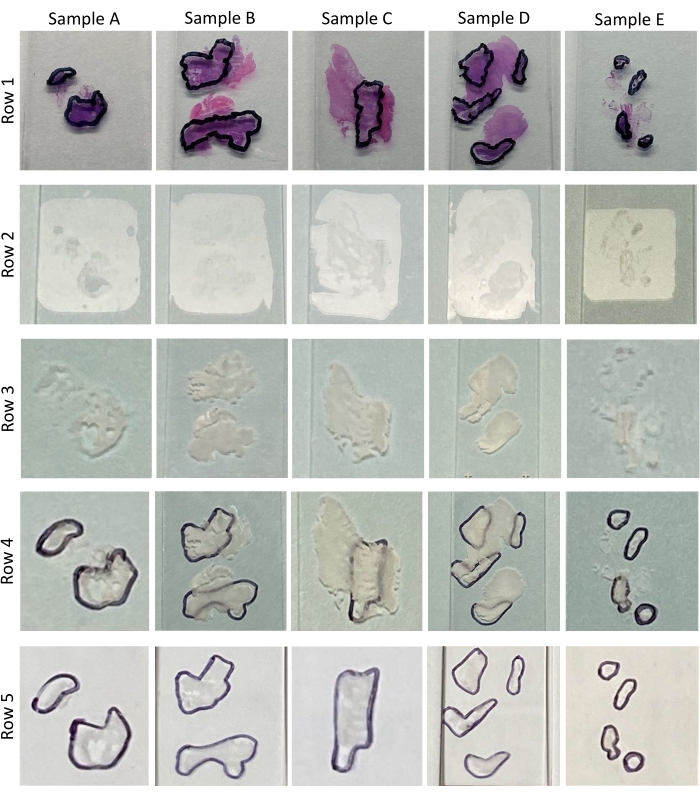
Figure 3: Tissue samples. This image demonstrates the pathologically reviewed and tumor marked H&Es (Row 1), unprocessed slide-mounted FFPE tissue sections (Row 2), deparaffinized slide-mounted FFPE tissue sections (Row 3), deparaffinized slide-mounted FFPE tissue sections with pathology markings traced on the back of the slide (Row 4), deparaffinized and macrodissected slide-mounted FFPE tissue sections (Row 5) for the 5 samples (A-E) used to demonstrate this protocol. Please click here to view a larger version of this figure.
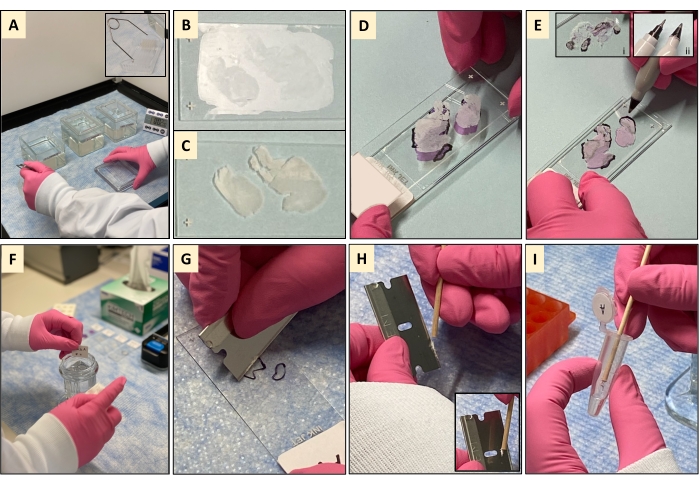
Figure 4: Deparaffinization and macro-dissection of FFPE tissue sections: (A) In a fume hood, FFPE tissues mounted in a slide rack are washed in two d-limonene washes and one ethanol wash. (B,C) Post wash, all the paraffin has been removed, and only the tissue remains on the slide, which is now white and highly visible compared to its prewashed counterpart. (D) The deparaffinized slide-mounted tissue section is placed face down on the back of its matching marked H&E. (E) The tumor area markings on the H&E are then traced on the back of the deparaffinized slide using a fine or ultrafine nibbed permanent marker (F) The marked deparaffinized slide-mounted tissue section is then dipped in a glycerol wash to dampen the tissue section for collection. The slide is removed slowly from the glycerol, and the back of the slide wiped with a tissue to remove excess glycerol before laying the slide tissue face-up on the bench. (G) Using the flat side of a clean razor blade, the tissue is macrodissected, and the unwanted tissue outside of the pathologist markings is discarded before collecting the tissue of interest, which gathers along the edge of the blade. (H) A wooden stick is used to remove the collected tissue from the edge of the blade. (I) The tissue is then transferred to a pre-labeled microtube prefilled tissue digestion buffer and is ready for nucleic acid extraction. Please click here to view a larger version of this figure.
| Sample ID | Whole Tissue (a) | Circled area of tissue (b) | Overall tumor content of whole tissue (c) | Fold increase in tumor content by macrodissection (d) | Not macrodissected (e) | Macrodissected (f) | ||||
| % Viable tumor | % Other | % Viable tumor | % Other | # of 5 µm slides extracted | RNA conc (ng/µL) | # of 5 µm slides extracted | RNA conc (ng/µL) | |||
| Sample A | 60 | 40 | 75 | 25 | 45 | 1.7 | 2 | 19.0 | 4 | 58.3 |
| Sample B | 60 | 40 | 65 | 35 | 39 | 1.7 | 1 | 34.0 | 2 | 60.0 |
| Sample C | 40 | 60 | 65 | 35 | 26 | 2.5 | 1 | 13.7 | 2 | 46.2 |
| Sample D | 35 | 65 | 90 | 10 | 32 | 2.9 | 1 | 57.3 | 2 | 60.0 |
| Sample E | 20 | 80 | 30 | 70 | 6 | 5.0 | 3 | 25.2 | 3 | 44.6 |
Table 1: Pathology review data. The table shows (a) the percentage of viable tumor in the whole tissue section by area, (b) the percentage of viable tumor in the area circled/marked by the pathologist during the review by cellularity, (c) the estimated overall tumor cellularity of the whole tissue (a x b), (d) the estimated fold increase in tumor cellularity achieved with macrodissection, (e and f) the number of 5 µm unstained FFPE slide-mounted tissue sections extracted and the resulting RNA concentrations for matched non-macrodissected and macrodissected samples. % Other refers to all other tissues present in a given sample that is not tumor tissue and can include connective tissue, stromal fibroblasts, blood vessels as well as other inherent stromal elements. Please click here to download this Table.
| Sample ID | RNA input (ng) | DLBCL90 COO call | DLBCL90 call probability | DHITsig call | DHITsig pos probability | DHITsig neg probability | |
| Not macrodissected | Sample A | 95.0 | GCB | 0.000 | NEG | 0.135 | 0.865 |
| Sample B | 170.0 | GCB | 0.000 | NEG | 0.032 | 0.968 | |
| Sample C | 68.5 | GCB | 0.028 | NEG | 0.033 | 0.967 | |
| Sample D | 286.5 | ABC | 0.998 | NEG | 0.002 | 0.998 | |
| Sample E | 126.0 | ABC | 0.989 | UNCLASS | 0.212 | 0.788 | |
| Macrodissected | Sample A_M | 291.7 | GCB | 0.000 | UNCLASS | 0.224 | 0.776 |
| Sample B_M | 300.0 | GCB | 0.000 | NEG | 0.016 | 0.984 | |
| Sample C_M | 231.2 | UNCLASS | 0.210 | NEG | 0.015 | 0.985 | |
| Sample D_M | 300.0 | ABC | 0.999 | NEG | 0.002 | 0.998 | |
| Sample E_M | 223.2 | ABC | 0.987 | NEG | 0.151 | 0.849 | |
| Macrodissected & RNA diluted | Sample A_M | 95.0 | GCB | 0.000 | UNCLASS | 0.254 | 0.746 |
| Sample B_M | 170.0 | GCB | 0.000 | NEG | 0.023 | 0.977 | |
| Sample C_M | 68.5 | UNCLASS | 0.117 | NEG | 0.027 | 0.973 | |
| Sample D_M | 286.5 | ABC | 0.999 | NEG | 0.002 | 0.998 | |
| Sample E_M | 126.0 | ABC | 0.995 | NEG | 0.167 | 0.833 | |
Table 2: DLBCL90 digital gene expression assay results. Five samples (A-E) were either not macrodissected or macrodissected before nucleic acid extractions were performed. The resulting RNA was run on the DLBCL90 assay, for which the maximum RNA input volume is 5 µL. Non-macrodissected samples were run using 5 µL of stock RNA. Each macrodissected sample was run twice using (a) 5 µL of stock RNA unless aliquots of 60 ng/µL were possible and (b) 5 µL of stock RNA diluted to match the concentrations/input of their non-macrodissected counterparts. The suffix _M denotes that that sample was macrodissected. Please click here to download this Table.
Discussion
FFPE tissues are frequently heterogeneous admixtures of tumor and non-tumor tissues. High sensitivity genomic tests are becoming increasingly prevalent in both clinical and research settings but can be confounded by the presence of contaminating non-tumor tissue. Indeed, a minimum tumor content of 60% is frequently recommended for genomic studies. Percentage tumor can be determined by the area of tissue occupied by the tumor material or by the proportion of tumor cells within the tissue. Although tumor by area is a commonly used metric for tumor purity, it does not always portray an accurate description of the tissue. Consider two tissues, both with 1000 cells, of which 500 are tumor cells. In tissue A, the 500 non-tumor cells are stromal cells with similar volumes to that of the tumor cell. In this tissue, the percentage of tumor can be considered 50% by both cellularity and area. In tissue B, the 500 non-tumor cells are fat cells with volumes that are 4x that of the tumor cell. In this tissue, the percentage tumor is still 50% by cellularity but 20% by area. A third tissue, tissue C, is composed of 500 tumor cells plus 400 fat cells and 800 stromal cells with volumes that are 4x and 0.5x that of the tumor cells, respectively. Given 100 fat cells equals the volume of 800 stromal cells, the percentage tumor of tissue C is 29% by cellularity (500/1700) but still 20% by area. Tissue D is also comprised of the tumor, fat, and stromal cells with volume ratios of 1x, 4x, and 0.1x. However, the number of cells is 400, 10, and 720, respectively. Thus, the percentage tumor of tissue D is 35% by cellularity (400/1130) but 78% by area. These examples are overly simplistic and do not reflect real-world tissue compositions but clearly convey the importance of tissue composition and the difference between tumor content by area and by cellularity. Importantly, when it comes to enriching tumor content for downstream nucleic acid extraction, tumor cellularity is the more important attribute due to the heightened confounding potential of extracting genomic material from more non-tumor cells than tumor cells. This not only underscores the need to assess the tumor content of tissues in terms of percentage cellularity but also the need to excise unwanted tissue in order to minimize any potential negative effects of the non-tumor tissue. There are several methods available for tissue enrichment, with the main ones being macrodissection and microdissection.
Macrodissection, the method described in this protocol, is relatively quick, simple, and does not require costly or specialized equipment. Although macrodissection can greatly improve tumor content, it is important to understand that it does not completely eliminate non-tumor material. The purpose of macrodissection is to enrich the tissue of interest sufficiently through the exclusion of unwanted tissue in order to reduce the "noise" stemming from unwanted tissue, which in turn can enhance the signal of interest from the tissue of interest. Thus, macrodissection mediated tumor enrichment is a way to enhance the signal-to-noise ratio in order to better detect markers of interest, particularly tumor-specific molecular markers with low abundance or poor expression. However, macrodissection has limitations due to the lack of precision offered by coarse tools such as razor blades and is susceptible to precision issues stemming from the line thickness of the pathologist's marker, as well as potential errors when tracing the pathologists H&E demarcations. As alluded above, it is not possible to achieve 100% tumor purity due to the presence of inherent and tumor-inducing stromal elements (i.e., connective tissue, stromal fibroblasts, blood vessels, benign reactive lymphocytes, macrophages) embedded within the tumor itself. Indeed, many invasive or diffusely infiltrative malignancies induce a robust desmoplastic stromal response, resulting in clusters of tumor cells that are intimately admixed with stromal fibroblasts and other non-neoplastic cell types; where tumors associated with this stromal reaction pattern, such as pancreatic cancer tissues21, may benefit more from digitally guided microdissection rather than manual macrodissection.
Manual microdissection is performed under a microscope to aid identification, dissection, and isolation of tissues specific cells or populations using a needle or scalpel and has the advantage of increased precision over macrodissection22. However, manual microdissection is a laborious process that lacks the finesse needed for complex tissues with low tumor contents or intricate features that are incompatible with manual dissection. Such tissues can be dissected using high-precision automated methods like laser capture microdissection. Indeed, digitally guided microdissection has been shown to yield higher percentage tumor contents compared to manual macrodissection in pancreatic cancer tissues23. However, the drawbacks of these high precision automated methods, such as the need for specialized, expensive equipment and highly trained individuals, have hindered its incorporation into workflows. A study by de Bruin et al. comparing the effects of macrodissection and laser capture microdissection (LCM) on gene expression profiling found that LCM samples had low total RNA yields (30 ng average) and required two rounds of mRNA amplification in order to meet the cDNA library prep input threshold24. The authors found that the resulting LCM gene expression profiles were affected by the rounds of mRNA amplification more than macrodissected profiles were affected by non-tumor stromal contributions and concluded that macrodissection could be adequately used to generate reliable gene expression data24.
A significant advantage of NanoString digital gene expression profiling, particularly when working with highly degraded FFPE derived RNA, is that it does not require enzymatic dependent processes such as RNA amplification or preparation of cDNA libraries. However, assays are typically optimized for inputs between 50-300 ng of total RNA25,26, which, based on the findings of de Bruin et al.24, may not be compatible with microdissected tissues without increasing the tissue input; an unfavorable demand in an era where tissue samples are increasingly collected as small biopsies rather than surgical resections. The RNA inputs used for the DLBCL90 assay ranged from 68.5-300 ng for both the macrodissected and non-dissected tissues. The results show that macrodissection resulted in call changes in 60% of the samples examined and that these changes were observed irrespective of RNA input of the macrodissected samples. However, the COO probability for the low RNA input did encroach the COO GCB/UNC probability call threshold, where the thresholds are 0 to <0.1 for GCB, 0.1-0.9 for UNC, and >0.9 to 1.0 for ABC calls20. The principal DLBCL COO subtypes are GCB and ABC, which make up 41% and 44% of all DLBCL cases, with UNC representing an intermediate group of the two and ABC being the most aggressive20,27. Thus, while the COO call change upon macrodissection of sample C did not cause a frank change in COO subtype from GCB to ABC, the change from GCB to UNC may suggest a shift towards a more aggressive disease. Moreover, recent studies indicate that the UNC subtype is not simply just an intermediate subtype and that it may potentially possess subtype-specific therapeutically exploitable attributes28. Similarly, macrodissection of samples A and E did not cause frank changes in DHITsig calls from DH negative to DH positive, or vice versa. However, the movements of a GCB sample (sample A) from NEG to UNCLASS and an ABC sample (sample E) from UNCLASS to NEG upon macrodissection are biologically appropriate as double hit translocations involving BCL2 are reported to be an exclusively GCB phenomenon19. Although translocations are traditionally and ubiquitously detected by FISH in clinical settings, there is a growing momentum to identify an alternative less involved and time-consuming method for their detection. The DLBCL90 assay is an important tool that addresses this need, where the rationale for its use is strengthened by the finding that this assay is capable of detecting translocations cryptic to FISH probes used in clinical diagnostics29.
The macrodissection protocol described above outlines a simple method that enables researchers to increase the tumor content of tissue samples that would ordinarily fall below commonly used study inclusion criteria thresholds. Including macrodissection in a study workflow enables researchers to salvage poorly tumor-dense tissues from study exclusion by increasing their tumor content. In turn, this permits increased confidence that the resulting RNA and DNA eluates represent the tumor under genomic investigation. Although other more precise methods for tissue dissection do exist, for tumors that grow in a more expansive, non-infiltrative, sheet-like, or solid fashion, macrodissection is likely sufficient. The results presented here highlight the importance of tumor purity in genomic assays and macrodissection as a reliable tool to achieve this.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is supported by NIH-funded AIDS and Cancer Specimen Resource (ACSR, UM1 CA181255-2) under its biospecimen science program. The video was filmed and post-production editing was performed by Mayo Clinic Media Services.
Materials
| Name | Company | Catalog Number | Comments |
| 200-proof ethanol | Decon | 2701 | |
| AllPrep DNA/RNA FFPE Kit | Qiagen | 80234 | DNA/RNA FFPE extraction kit |
| Coplin pots | Various | x | |
| DLBCL90 probes | NanoString | various | Digital gene expression profiling based DLBCL90 assay |
| d-Limonene | VWR | 89376-092 | |
| Forceps | Various | x | |
| Glass micrscope slides | FisherBrand | 12-550-15 | |
| Glycerol | VWR | 0854-1L | |
| Master kits | NanoString | various | |
| Microtome | Leica | RM2265 | |
| Microtubes | Ambion | AM12400 | |
| NanoDrop One | Thermo Scientific | ND-ONE-W | Spectrophotometer for DNA, RNA and protein qualitation |
| nCounter | NanoString | x | Digital gene expression profiling platform used to run the DLBCL90 assay |
| Permanent marker | Electrib Microscope Sciences | 72109-12 | |
| Razor blade dispenser | Electrib Microscope Sciences | 71985-10 | |
| Razor blades | Electrib Microscope Sciences | 71985-23 | |
| Tissue digestion buffer | Qiagen | 80234 | |
| Ultrapure water | VWR | SH30538.02 | |
| Waterbath | Triangle Biomedical Sciences | TFB-120 | |
| Wooden stick | FisherBrand | 22363158 |
References
- Mathieson, W., Thomas, G. A. Why formalin-fixed, paraffin-embedded biospecimens must be used in genomic medicine: An evidence-based review and conclusion. Journal of Histochemistry and Cytochemistry. 68 (8), 543-552 (2020).
- Robetorye, R. S., Maguire, A., Rosenthal, A. C., Rimsza, L. M. Profiling of lymphoma from formalin-fixed paraffin-embedded tissue. Seminars in Hematology. 56 (1), 46-51 (2019).
- Moorcraft, S. Y., Gonzalez, D., Walker, B. A. Understanding next generation sequencing in oncology: A guide for oncologists. Critical Reviews in Oncology/Hematology. 96 (3), 463-474 (2015).
- Haile, S., et al. Automated high throughput nucleic acid purification from formalin-fixed paraffin-embedded tissue samples for next generation sequence analysis. PLoS One. 12 (6), 0178706 (2017).
- Oh, E., et al. Comparison of accuracy of whole-exome sequencing with formalin-fixed paraffin-embedded and fresh frozen tissue samples. PLoS One. 10 (12), 0144162 (2015).
- Holley, T., et al. Deep clonal profiling of formalin fixed paraffin embedded clinical samples. PLoS One. 7 (11), 50586 (2012).
- . TCGA Tissue sample requirements: High quality requirements yield high quality data Available from: https://www.cancer.gov/about-nci/organization/ccg/structural-genomics/tcga/studied-cancers (2021)
- Javey, M., et al. Innovative tumor tissue dissection tool for molecular oncology diagnostics. The Journal of Molecular Diagnostics: JMD. 23 (4), 399-406 (2021).
- Feldman, A. T., Wolfe, D. Tissue processing and hematoxylin and eosin staining. Methods in Molecular Biology. 1180, 31-43 (2014).
- Duan, Q., Zhang, H., Zheng, J., Zhang, L. Turning cold into hot: Firing up the tumor microenvironment. Trends in Cancer. 6 (7), 605-618 (2020).
- Kim, Y. W., et al. Safety evaluation and risk assessment of d-Limonene. Journal of Toxicology and Environmental Health Part B: Critical Reviews. 16 (1), 17-38 (2013).
- Foti, C., et al. Occupational contact dermatitis to a limonene-based solvent in a histopathology technician. Contact Dermatitis. 56 (2), 109-112 (2007).
- Meuse, C. W., Barker, P. E. Quantitative infrared spectroscopy of formalin-fixed, paraffin-embedded tissue specimens: paraffin wax removal with organic solvents. Applied Immunohistochemistry and Molecular Morphology. 17 (6), 547-552 (2009).
- Schmeller, J., et al. Setting out the frame conditions for feasible use of FFPE derived RNA. Pathology - Research and Practice. 215 (2), 381-386 (2019).
- Prema, V., et al. Biofriendly substitutes for xylene in deparaffinization. Journal of Pharmacy and Bioallied Sciences. 12, 623-630 (2020).
- Ennishi, D., et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. Journal of Clinical Oncology. 37 (3), 190-201 (2019).
- Alizadeh, A. A., et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403 (6769), 503-511 (2000).
- Rosenwald, A., et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England Journal of Medicine. 346 (25), 1937-1947 (2002).
- Scott, D. W., et al. High-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements with diffuse large B-cell lymphoma morphology. Blood. 131 (18), 2060-2064 (2018).
- Scott, D. W., et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 123 (8), 1214-1217 (2014).
- Heinrich, M. A., Mostafa, A., Morton, J. P., Hawinkels, L., Prakash, J. Translating complexity and heterogeneity of pancreatic tumor: 3D in vitro to in vivo models. Advanced Drug Delivery Reviews. 174, 265-293 (2021).
- Erickson, H. S., Gillespie, J. W., Emmert-Buck, M. R. Tissue microdissection. Methods in Molecular Biology. 424, 433-448 (2008).
- Geiersbach, K., et al. Digitally guided microdissection aids somatic mutation detection in difficult to dissect tumors. Cancer Genetics. 209 (1-2), 42-49 (2016).
- de Bruin, E. C., et al. Macrodissection versus microdissection of rectal carcinoma: minor influence of stroma cells to tumor cell gene expression profiles. BMC Genomics. 6, 142 (2005).
- Ramsower, C. A., et al. Clinical laboratory validation of the MCL35 assay for molecular risk stratification of mantle cell lymphoma. Journal of Hematopathology. 13 (4), 231-238 (2020).
- Maguire, A., et al. Enhanced DNA repair and genomic stability identify a novel HIV-related diffuse large B-cell lymphoma signature. International Journal of Cancer. 145 (11), 3078-3088 (2019).
- Rosenwald, A., Staudt, L. M. Gene expression profiling of diffuse large B-cell lymphoma. Leukemia & Lymphoma. 44, 41-47 (2003).
- Wright, G. W., et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 37 (4), 551-568 (2020).
- Hilton, L. K., et al. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood. 134 (18), 1528-1532 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved