Method Article
Rapid, Cost-Efficient, Enzyme-Free Passaging of Human Pluripotent Stem Cells on Feeder Cells by Ethylenediaminetetraacetic Acid-Mediated Dis-Adhesion
In This Article
Summary
To avoid the limitations associated with the enzymatic or mechanical passaging of human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) cultured on feeder cells, we have established a fast, effective, cost-efficient, high-yield method for harvesting hESC or hiPSC colonies maintained on a feeder cell layer of human foreskin fibroblasts using EDTA-mediated dis-adhesion.
Abstract
Human pluripotent stem cells (human embryonic stem cells, hESCs, and human induced pluripotent stem cells, hiPSCs) were originally cultured on different types of feeder cells for maintenance in an undifferentiated state in long-term culture. This approach has been supplanted to a large extent by feeder-free culture protocols, but these involve more costly reagents and can promote a transition to a primed state, which restricts the cells' differentiation capacity. In both feeder and feeder-free conditions, the harvesting of hESC or hiPSC colonies for passaging is a necessary procedure for expanding the cultures.
To provide an easy and high-yield procedure for passaging hESCs/hiPSCs cultured on feeder cells, we have established a harvesting method using dis-adhesion elicited by the calcium chelator ethylenediaminetetraacetic acid (EDTA). We have assessed the yield and quality of the resultant passaged cells by comparing this approach to the original mechanical harvesting approach, in which colonies are isolated with a scalpel under a microscope (mechanical harvesting was chosen as a comparator to avoid the reagent variability associated with enzymatic harvesting).
In one set of experiments, two different hESC lines were maintained on a feeder cell layer of human foreskin fibroblasts. Each line was subjected to multiple passages using EDTA-based or mechanical harvesting and assessed for colony size and morphology, cell density, stemness marker expression, differentiation to the three germ layers in embryoid bodies, and genomic aberrations. In another set of experiments, we used EDTA-based harvesting on two different hiPSC lines and obtained similar results. EDTA-induced dis-adhesion saved time and gave a higher yield of colonies of a more favorable size and more uniform morphology compared to mechanical harvesting. It was also faster than enzymatic harvesting and not prone to enzyme batch variability. The EDTA-induced dis-adhesion method also facilitates the transfer of hESC/hiPSC lines from feeder cell-based culture to feeder-free conditions if desired for downstream use and analysis.
Introduction
The proper maintenance of hESCs and hiPSCs in vitro is a basic and convenient methodology for several avenues of research in human cell and developmental biology. Due to the inherent drive of hESCs and hiPSCs to differentiate, maintaining the undifferentiated state in vitro demands particular care and attention. Thus, developing cost-efficient protocols for the maintenance and passaging of hESCs and hiPSCs with as little methodological variability as possible is of great general utility.
Originally, hESCs and hiPSCs were cultured on different types of feeder cells to assist in the long-term culture and maintenance of the undifferentiated state1,2,3. More recently, culture under feeder-free conditions has become the norm, as it avoids dealing with feeder cells altogether4. However, some laboratories and core facilities still culture hESCs or hiPSCs on feeder cells. Feeder-free culture is more expensive because it requires the use of culture media of special compositions and some form of coating of the culture surface to ensure colony adherence (major extracellular matrix [ECM] components or a commercial ECM compound, or using commercially available coated plates). The expense is not trivial and presents a potential financial hindrance for some laboratories interested in pursuing hESC- or hiPSC-based research and development. Moreover, culture under feeder-free conditions tends to drive the hESCs and hiPSCs to a less naive state than is maintained on feeder cells5, and this can compromise subsequent differentiation and lead to genetic variations6.
Historically, the passaging of hESCs and hiPSCs cultured on feeder cells involved mechanical harvesting - using a scalpel to excise colonies under a microscope7 - but this was later largely supplanted by enzymatic digestion with or without gentle scraping to isolate colonies or dissociated cells. Mechanical harvesting is tedious and requires precision microsurgery. Enzymatic harvesting can vary in efficiency due to batch-to-batch enzyme differences and tends to favor complete dissociation, which promotes cell death unless counteracted by ROCK inhibitors8,9 and increases the incidence of abnormal karyotypes9.
To take advantage of the lower expense and greater differentiation potential of culturing hESCs and hiPSCs on feeder cells while avoiding the disadvantages of mechanical and enzymatic harvesting, we have established a fast, effective, cost-efficient, high-yield method for harvesting hESC and hiPSC colonies maintained on a feeder layer of human foreskin fibroblasts using EDTA-mediated dis-adhesion. We have compared the yield, variability, and stem cell quality to that obtained with mechanical harvesting (we did not compare to enzymatic digestion because of the additional variability this approach entails). We note that EDTA-mediated dis-adhesion also works well for transferring colonies from feeder-based culture to feeder-free conditions, if desired for downstream use and analyses. This method provides a transition with a consistent passaging method, since EDTA-induced dis-adhesion is a popular approach employed for feeder-free cultures.
Protocol
See the Table of Materials for details about all the materials, reagents, and instruments used in this protocol.
1. Cultivation of human fibroblast cells and preparation of the feeder cell layer
- Seed 0.5 × 106 human foreskin fibroblasts (hereafter called "feeder cells") per each T-75 culture flask (number of flasks as needed) with 20 mL of Iscove's Modified Dulbecco's Medium (IMDM) with (w/) 10% fetal bovine serum (FBS), hereafter called "feeder cell medium."
- When the feeder cells reach 90% confluence, remove the medium, and wash 3x with 10 mL of Dulbecco's Phosphate-Buffered Saline (DPBS) per flask to avoid the inhibition of trypsin by factors in the medium. Add 2 mL of trypsin-EDTA to each flask, and place the flask(s) in a 37 °C/5% CO2 incubator for 5 min or until the feeder cells have detached from the flask(s). Observe the detachment of the cells under a microscope as floating aggregates of cells or single cells.
- Add 5 mL of fresh pre-warmed feeder cell medium to each flask to inactivate the trypsin-EDTA, and gently suspend the feeder cells by pipetting.
- Transfer the feeder cells to a 15 mL centrifuge tube. Cap the tube, and pellet the feeder cells by centrifugation at 200 × g for 5 min.
- Carefully remove the supernatant without disturbing the feeder cell pellet. Then, carefully resuspend the pellet in 4 mL of fresh feeder cell medium. Ensure that the feeder cells are thoroughly resuspended before counting using a cell counting chamber or other cell counting apparatus.
- Add 0.5 × 106 feeder cells to the required number of new T-75 culture flasks for expansion, and add 20 mL of fresh feeder cell medium to each flask. Incubate the culture flask(s) in a 37 °C/5% CO2 incubator until the feeder cells have reached 90% confluence.
NOTE: Feeder cells can be used up to at least passage 25. - Calculate the number of feeder cells required for the number of 35 mm tissue culture dishes that will be used for culturing the hESCs/hiPSCs.
NOTE: Usually, 3.0 × 105 feeder cells per tissue culture dish are sufficient to generate a confluent layer of feeder cells. - To avoid the proliferation of the feeder cells, ensure that they are mitotically arrested in either of two ways.
NOTE: For both methods, a large batch of mitotically arrested feeder cells can be generated and frozen down in aliquots for later use.- Perform mitotic arrest by gamma-irradiation by transferring all the feeder cells needed to a 50 mL centrifuge tube and topping up with feeder cell medium to a total volume of 5 mL. Transport immediately at room temperature to a gamma-irradiation machine, and irradiate to mitotically arrest the feeder cells (300 kV and 10 mA for 20 min).
NOTE: A delay in transport can lead to undesired attachment of the feeder cells to the wall of the 50 mL centrifuge tube. If the transport requires more than a few minutes, ensure that the feeder cells remain suspended during transport by continuously agitating the tube. - Perform mitotic arrest using mitomycin C by transferring all the feeder cells needed in 5 mL of feeder cell medium to a 50 mL centrifuge tube, and then add 15 mL of feeder cell medium containing 20 μg/mL mitomycin C, and incubate in a 37 °C/5% CO2 incubator for 3 h. Add 20 mL of 37 °C PBS, pellet the cells by centrifugation at 200 × g for 5 min, repeat the PBS wash two additional times, and resuspend in feeder cell medium.
- Perform mitotic arrest by gamma-irradiation by transferring all the feeder cells needed to a 50 mL centrifuge tube and topping up with feeder cell medium to a total volume of 5 mL. Transport immediately at room temperature to a gamma-irradiation machine, and irradiate to mitotically arrest the feeder cells (300 kV and 10 mA for 20 min).
- After the feeder cells have been mitotically arrested, return to the tissue culture hood, and plate out the feeder cells at 3.0 × 105 cells per 35 mm tissue culture dish, as follows. Ensure that the feeder cells are fully resuspended, add feeder cell medium to reach a feeder cell concentration of 1.5 × 105 per mL, and add 2 mL of this feeder cell suspension to each 35 mm tissue culture dish.
- Transfer the culture dishes to a 37 °C/5% CO2 incubator. To ensure an even distribution of the feeder cells, move the culture dishes slowly but firmly on the incubator shelf forward and backward 3x, followed by a pause, and then perform the same action from left to right 3x. Do not move the dishes again, and gently close the incubator door.
- After 24 h, switch from feeder cell medium to IMDM w/ 10% serum replacement (SR). Replace this medium thereafter every third day. The feeder cells are ready for use after the first 3 days.
2. Mechanical harvesting of the hESC or hiPSC colonies
- Prewarm hESC medium consisting of 80% Dulbecco's Modified Eagle's Medium (DMEM), 20% SR, 1 mM glutamine substitute 100x, 1 mM non-essential amino acids (NEAA), 1 mM penicillin/streptomycin (P/S), 0.1 mM 2-mercaptoethanol, and 10 ng/mL basic fibroblast growth factor (bFGF). The hESC medium is used for the culture of either the hESCs or hiPSCs on the feeder cells.
- Take fresh 35 mm tissue culture dishes containing mitotically arrested feeder cells, and replace the feeder cell medium with 1.2 mL of hESC medium containing bFGF at least 30 min before the transfer of the hESC/hiPSC colonies.
- Place a culture dish containing hESC/hiPSC colonies on mitotically arrested feeder cells under a microscope with 10x magnification placed within a laminar flow hood. Use a sterile scalpel to cut carefully around the circumference of each colony and then cut each colony into 5-6 roughly equal pieces. Carefully lift up the colony pieces with the tip of the scalpel blade so that they detach from the feeder cell layer and float freely in the medium.
- Try to avoid regions of the colonies that contain differentiating cells, which appear as islands of smaller cells with less distinct nuclei compared to hESCs/hiPSCs within a colony.
- Transfer the freely floating colonies with a 1 mL pipette to the new culture dishes containing the feeder cells. Try to keep the colonies separate so that they do not grow into each other later. Move the culture dishes carefully to a cell incubator, and avoid disturbing the dishes until the next day.
- On the following day, carefully add 600 µL of hESC medium containing bFGF to a final volume of 1.8 mL. Replace the hESC + bFGF medium each day thereafter until the next passage (generally after 1 week).
3. Harvesting of the hESC or hiPSC colonies using EDTA-mediated dis-adhesion
- Take fresh culture dishes with mitotically arrested feeder cells, and switch from IMDM w/ 10% SR to 1.2 mL of prewarmed hESC + bFGF medium at least 30 min before the transfer of the colonies.
- Handle one culture dish containing hESC or hiPSC colonies at a time. Remove the hESC + bFGF medium, and wash the colonies with 1 mL of room-temperature DPBS to eliminate any possible unattached cells and cell debris. Add 1 mL of 0.5 mM EDTA, and incubate for 1 min at 37 °C. If the laminar flow hood has a warming plate, perform this step and the steps in section 4 on the warming plate for better dis-adhesion.
- After the 1 min incubation, remove the EDTA solution, and carefully add 1 mL of hESC + bFGF medium using a 1 mL pipette. Gently triturate with the same pipette to release the colonies from the feeder cell layer. Continue to triturate carefully until the feeder cell layer loosens and folds on itself in a separate clump. Pull away the feeder cell layer with the pipette tip.
- Transfer the suspended hESC/hiPSC colonies with a new 1 mL pipette to new culture dishes containing the feeder cells and hESC + bFGF medium, splitting at a ratio of 1:5. The colonies tend to distribute evenly within each new culture dish, but facilitate this by moving the dish gently from side to side. Replace the hESC + bFGF medium each day thereafter until the next passage (generally after 1 week).
Results
In the assays and comparisons documented below, we used two hESC lines (H9 and HS429, from WiCell and the Karolinska Institute, respectively) and two hiPSC lines (NCS001 and NCS002, both generated by the Norwegian Core Facility for Human Pluripotent Stem Cells). The data presented in the figures and tables are from the hESC lines, but entirely similar results were obtained from the hiPSC lines.
In our hands, mechanical harvesting resulted in the colonies being split into approximately five to six clumps of ~200-250 µm in diameter, whereas with EDTA-induced dis-adhesion followed by trituration, each colony was split into ~10-20 clumps of ~60 µm in diameter. We estimate that the number of cells in each EDTA-harvested clump is ~20. As it is impractical to split a colony into clumps of this size with a scalpel, in this respect, EDTA-induced dis-adhesion is superior, as it generates clumps of a size that is more favorable for colony cell survival10,11.
The hESC/hiPSC colonies harvested using EDTA were also more homogeneous in size and shape compared to the colonies harvested mechanically (Figure 1A-F). This is because the cutting required for mechanical harvesting generates uneven edges and varying clump sizes. To evaluate this quantitatively, we assessed the colony circularity (as a measure of how rounded the colony edges were; a value of 1 indicates a perfect circle) 5 days after passaging using the ImageJ-win64 protocol12. The colony circularity was significantly lower in the colonies harvested mechanically (mechanical harvesting: 0.61 ± 0.10; EDTA-based harvesting: 0.84 ± 0.01; n = 10, p < 0.001, Mann-Whitney U-test, U = 10).
The cell density in the harvested and replated colonies, which is a measure of post-harvesting cell-cell interactions during colony formation, was similar with EDTA-based harvesting and mechanical harvesting (Table 1 and Figure 1G,H). The mechanically harvested colonies had a greater tendency to develop necrosis in their central regions (Figure 1J). This was likely due to variability in the shape and, particularly, the size of the mechanically isolated cell clumps, as when these clumps are too large, they can easily fold upon themselves when being transferred to new culture dishes. This was not the case with the colonies harvested using EDTA, which uniformly exhibited a translucent appearance with distinct edges (Figure 1I).
Using EDTA-based harvesting, we were able to collect essentially all the colonies that had been established in a well within 2-3 min. Using mechanical harvesting, collecting all the colonies in a well would be tedious and time-consuming. We typically managed to collect only ~30%, or ~20-25 colonies, using mechanical harvesting, and this took ~20 min. Likewise, using collagenase digestion followed by gentle scraping, it was typically difficult to harvest all the colonies, although the total procedure only took a few minutes. Thus, EDTA-based harvesting is as fast or faster than enzymatic harvesting and more efficient than either mechanical or enzymatic harvesting.
To assess the effect of the different harvesting methods on stemness and pluripotency, we first subjected the colonies obtained after 20 passages using EDTA-based or mechanical harvesting to qPCR analysis (Figure 2) and immunocytochemical staining (Figure 3 and Figure 4) for stemness markers. The colonies obtained using either method exhibited a stable expression of stemness markers both at the mRNA and protein levels. We then assessed pluripotency by differentiation to the three germ layers in embryoid bodies (Figure 5 and Figure 6). The embryoid bodies generated from the hESCs or hiPSCs obtained after 20 passages using either method contained a mixture of cells expressing commonly assessed markers for ectoderm, mesoderm, and endoderm.
Finally, we assessed the incidence of genomic aberrations in hESCs and hiPSCs passaged by each method using qPCR-based genetic analysis (see the Table of Materials). The colonies obtained after 20 passages using either harvesting method exhibited some examples of modest deviation from a reference diploid chromosomal pattern (the abnormalities assessed were those commonly associated with the reprogramming of hiPSCs but can also be obtained in hESCs) (Figure 7). However, the pattern of these deviations was essentially the same in the colonies obtained after either harvesting method, indicating that they were not linked to the harvesting method.
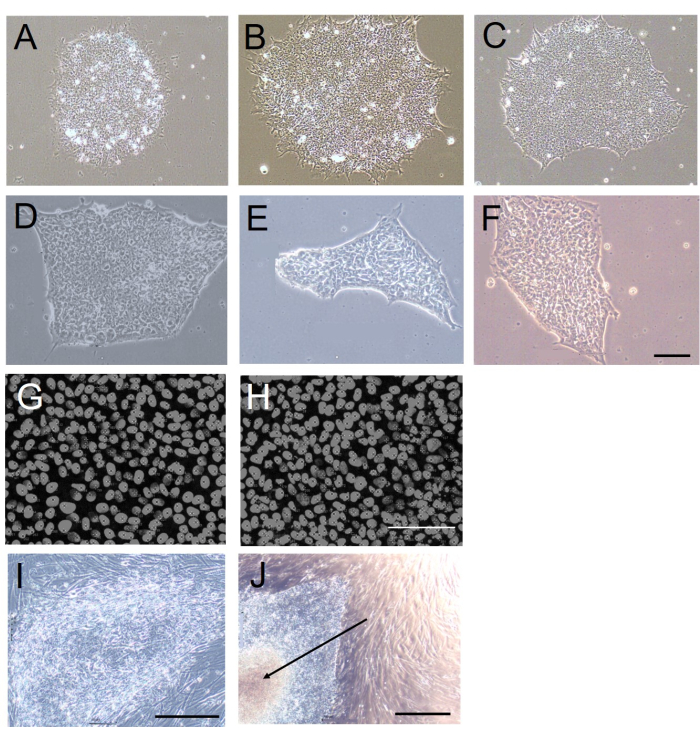
Figure 1: Colony morphology and cell density following EDTA-based or mechanical harvesting. (A-F) Representative brightfield images of H9 hESC colonies established in feeder-free culture for 5 days after 20 passages using (A-C) EDTA-based or (D-F) mechanical harvesting. (G,H) Representative fluorescence images of the cell density in H9 hESC colonies established after 20 passages using (G) EDTA-based or (H) mechanical harvesting. The cell nuclei are stained with DAPI. (I,J) Representative brightfield images of H9 hESC colonies established after 20 passages using (I) EDTA-based or (J) mechanical harvesting. Note the necrotic central region in the colony harvested mechanically (arrow in J). All the images were acquired 5 days following the 20th passage. Scale bars = 100 µm. Abbreviations: hESC = human embryonic stem cell; EDTA = ethylenediaminetetraacetic acid; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
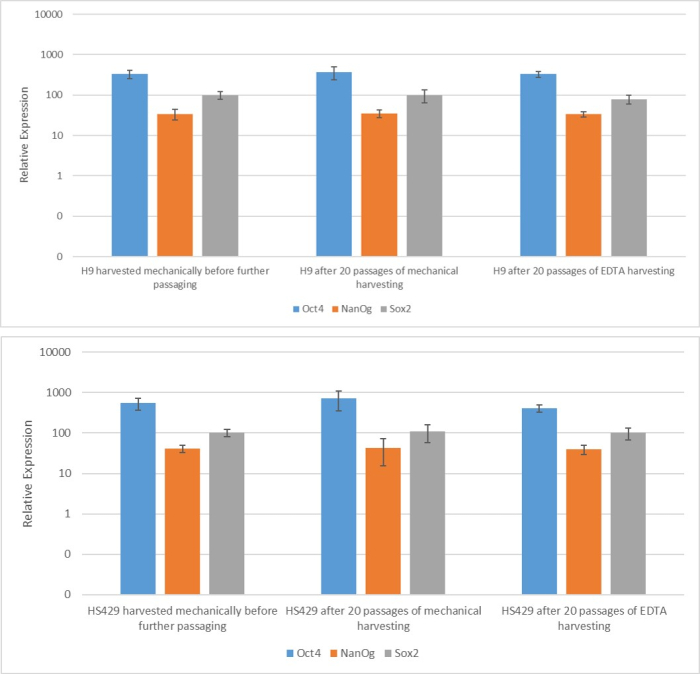
Figure 2: Expression of stemness marker mRNA in two hESC lines (H9 and HS429) generated after EDTA-based or mechanical harvesting. Quantitative real-time polymerase chain reaction of the indicated markers in H9 (upper panel) and HS429 (lower panel) hESCs after a single passage using mechanical harvesting, after 20 passages using mechanical harvesting, and after 20 passages using EDTA-based harvesting (1:5 dilution). The expression level is relative to that of the housekeeping gene ACTB (beta-actin). The error bars indicate the standard deviation. Abbreviations: hESC = human embryonic stem cell; EDTA = ethylenediaminetetraacetic acid. Please click here to view a larger version of this figure.
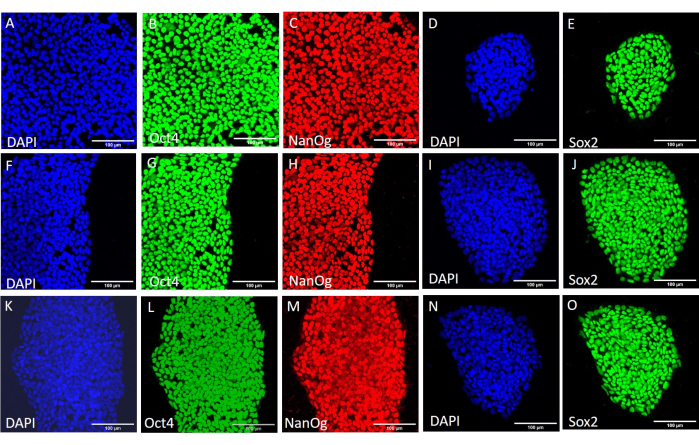
Figure 3: Expression of stemness marker proteins in the H9 hESC line after different harvesting conditions. Representative immunofluorescence staining of H9 hESC colonies harvested mechanically (A-E) before further passaging, (F-J) after 20 passages using mechanical harvesting, and (K-O) after 20 passages using EDTA-based harvesting. Scale bars = 100 µm. Abbreviations: hESC = human embryonic stem cell; EDTA = ethylenediaminetetraacetic acid; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
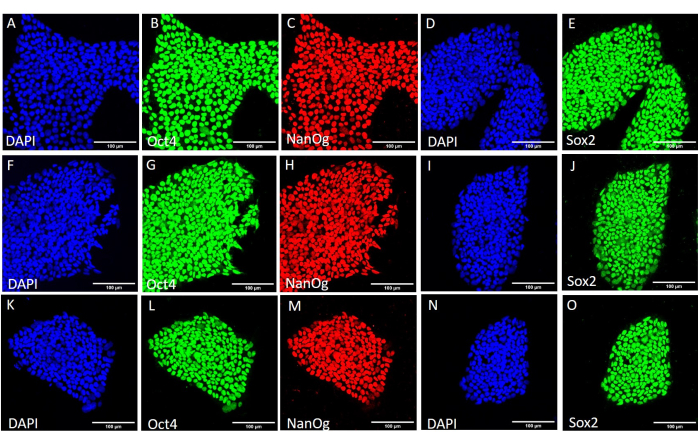
Figure 4: Expression of stemness marker proteins in the HS429 hESC line after different harvesting conditions. Representative immunofluorescence staining of HS429 hESC colonies harvested mechanically (A-E) before further passaging, (F-J) after 20 passages using mechanical harvesting, and (K-O) after 20 passages using EDTA-based harvesting. Scale bars = 100 µm. Abbreviations: hESC = human embryonic stem cell; EDTA = ethylenediaminetetraacetic acid; DAPI = 4',6-diamidino-2-phenylindole. Please click here to view a larger version of this figure.
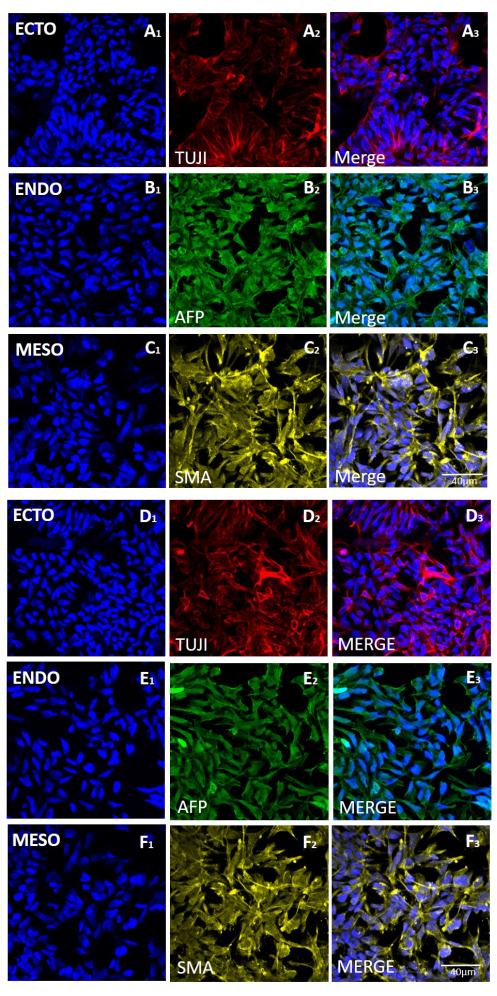
Figure 5: Expression of markers for the three germ layers in embryoid bodies generated from the H9 hESC line following mechanical or EDTA-based harvesting. Representative immunofluorescence staining of markers for (A and D rows) ectoderm (ECTO, TUJI), (B and E rows) endoderm (ENDO, AFP), and (C and F rows) mesoderm (MESO, SMA). EBs generated (A-C) after 20 passages of mechanical harvesting or (D-F) after 20 passages of EDTA-based harvesting. Scale bars = 40 µm. Abbreviations: hESC = human embryonic stem cell; EDTA = ethylenediaminetetraacetic acid; EBs = embryoid bodies. Please click here to view a larger version of this figure.
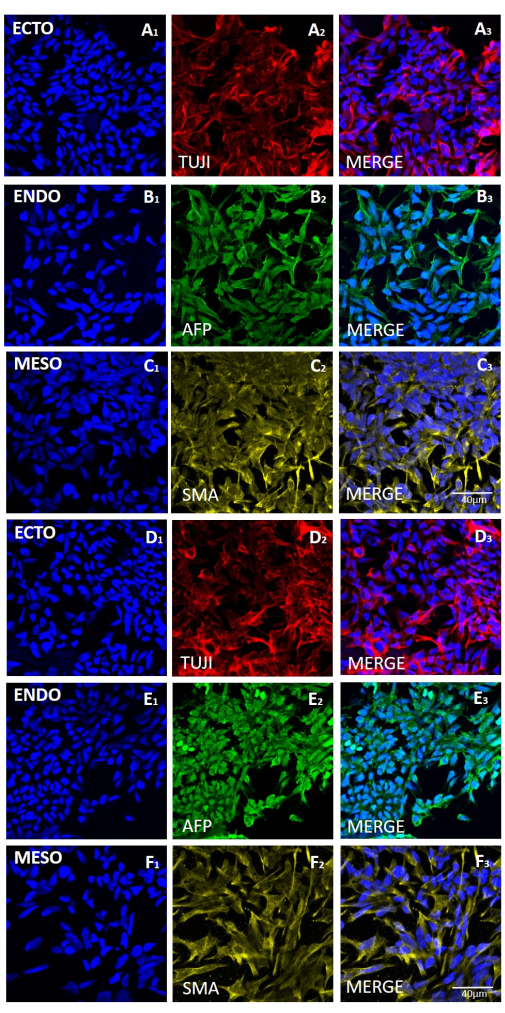
Figure 6: Expression of markers for the three germ layers in embryoid bodies generated from the HS429 hESC line following mechanical or EDTA-based harvesting. Representative immunofluorescence staining of markers for (A and D rows) ectoderm (ECTO, TUJI), (B and E rows) endoderm (ENDO, AFP), and (C and F rows) mesoderm (MESO, SMA). EBs generated (A-C) after 20 passages of mechanical harvesting (A-C) or (D-F) after 20 passages of EDTA-based harvesting. Scale bars = 40 µm. Abbreviations: hESC = human embryonic stem cell; EDTA = ethylenediaminetetraacetic acid; EBs = embryoid bodies. Please click here to view a larger version of this figure.
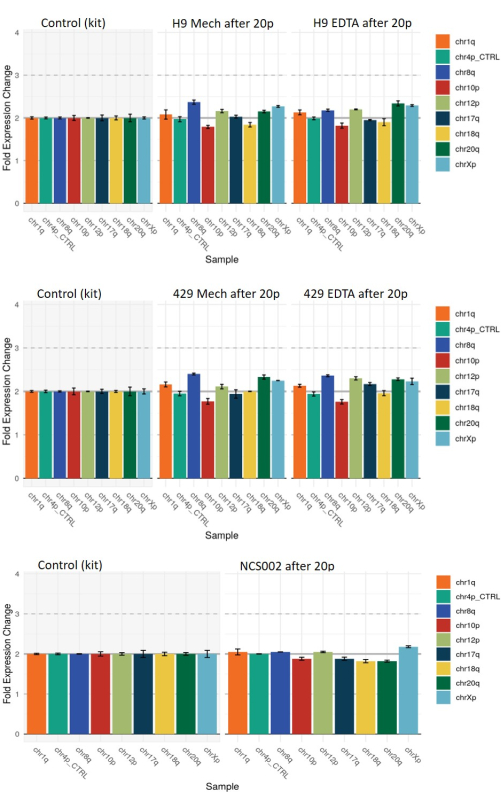
Figure 7: qPCR-based genetic analysis of common genomic aberrations in the HS9 and HS429 ESC lines and the NCS002 iPSC line following 20 passages using mechanical or EDTA-based harvesting. The baseline at value 2 represents normal diploidy at all the chromosomal markers. A value of 1 or 3 would represent a loss or gain, respectively, of the indicated chromosomal marker in all the cells. Intermediate values between 1 and 2 or between 2 and 3 indicate the presence of a loss or gain of the indicated marker in a fraction of the cells. Note that the pattern of aberrations is similar in the two harvesting conditions. Abbreviations: ESC = embryonic stem cell; EDTA = ethylenediaminetetraacetic acid; iPSC = induced pluripotent stem cell. Please click here to view a larger version of this figure.
| Cell density (cells/mm2) | ||
| H9 | mean | stdev |
| Mechanical harvest before further passaging | 3918 | 263.3 |
| Mechanical harvest 20 times | 3868 | 197.7 |
| EDTA harvest 20 times | 4080 | 127.8 |
| HS429 | mean | stdev |
| Mechanical harvest before further passaging | 5249 | 565.4 |
| Mechanical harvest 20 times | 5247 | 726.3 |
| EDTA harvest 20 times | 4963 | 448.8 |
Table 1: Comparison of the cell densities in the colonies from the two hESC lines (H9 and HS429) generated after EDTA-based or mechanical harvesting. The cell densities were assessed either after a single passage using mechanical harvesting, after 20 passages using mechanical harvesting, or after 20 passages using EDTA-based harvesting (at 1:5 dilution). In all cases, n = 5 colonies.
Discussion
We have described a rapid and cost-efficient method for harvesting hESCs and hiPSCs cultured on feeder cells using EDTA-mediated dis-adhesion and compared this primarily to the conventional method of mechanical harvesting using a scalpel. We also compared EDTA-based harvesting to enzymatic harvesting with respect to the speed of the method but not aspects of the resultant colony quality. The reason for this is that enzymatic harvesting is inherently more variable and has been linked to a higher prevalence of genomic aberrations5, which could obscure the inter-method differences.
We demonstrate that EDTA-based harvesting is faster and more efficient than either of the other methods and generates smaller and morphologically more homogeneous colonies than mechanical harvesting. This latter feature is beneficial with respect to cell survival, since the larger clumps obtained with mechanical harvesting are prone to central necrosis, while enzymatic digestion tends to generate isolated hESCs and hiPSCs, which are more prone to apoptosis and require extra treatment, for example with ROCK inhibitors, to survive. EDTA-based harvesting can be used for at least 20 passages. The EDTA-based and mechanical harvesting methods are comparable when it comes to colony cell density, mRNA and protein expression of stemness genes, differentiation of the three germ layers in embryoid bodies, and genomic abnormalities. If the goal is efficiency, higher yield, less variability, and gentler handling of the hESCs and hiPSCs, EDTA-based harvesting is preferable.
We also note that EDTA-based harvesting of hESCs and hiPSCs cultured on feeder cells is an inexpensive way to maintain a more naive state and provides a smooth transition from feeder-based to feeder-free culture where this is desirable.
Critical steps within the protocol
The most critical steps of EDTA-mediated dis-adhesion are protocol section 3 (incubation in the EDTA solution) and section 4 (trituration). If the exposure to the EDTA solution is longer than 1 min, the risk of complete dissociation to single cells increases. This can also occur if the trituration is too protracted or too harsh. The latter is affected by the pipette tip size. Using 1 mL cell culture pipettes as described here is ideal. Using a different type of pipette with a smaller tip diameter is risky.
Troubleshooting
If the feeder cells continue to proliferate, mitotic arrest has not been effective, and a fresh batch must be taken and the procedure restarted. If the colonies do not loosen from the feeder cell layer, one must make sure there is no Ca2+ in the EDTA and that the culture dish containing the colonies is rinsed well with PBS to remove any remaining cell culture medium before adding the EDTA. Too much dissociation, which generates isolated cells or cell clumps that are too small, can arise due to excessive trituration and compromises the establishment of new colonies. The degree of trituration should be determined empirically in trial runs of the protocol to confirm that the resultant cell clumps are ~60 µm in diameter. If the feeder layer detaches spontaneously from the culture dish, especially before the hESCs/hiPSCs are ready for harvesting, it could be because the feeder cells have not been used within ~7 days after being prepared. Therefore, the time frame of use of the feeder cells should be monitored carefully. If the feeder layer dissociates during EDTA exposure (something we have never observed with the feeder cells used here), either the type of feeder cell or their culture method must be changed.
Limitations of the technique
The main limitation of the technique is that it requires visual inspection of the dis-adhesion process to achieve a successful result. This means that users must learn how to identify when the colonies release from the feeder cell layer and the feeder cell layer loosens from the substrate. However, this is not difficult, and in our experience, new users of the technique can master it within a couple of trials.
There is also an inherent possibility that the harvested hESCs or hiPSCs may be contaminated by a few feeder cells. If the intention is to transfer to non-feeder conditions or to isolate the hESCs or hiPSCs for assays, such contamination would compromise purity. We note that with the feeder cells used here (human foreskin fibroblasts), it is extremely difficult to dissociate the feeder cell layer, even with enzymatic digestion (not shown). Since the undissociated feeder cell layer is removed in toto, contamination of the harvested hESCs or hiPSCs is likely to be negligible. As the feeder cells are, moreover, mitotically arrested, any contamination would eventually diminish to nil with further passaging of the hESCs or hiPSCs.
Significance with respect to existing methods
The current norm for culturing hESCs and hiPSCs is to do so under feeder-free conditions, for which the use of EDTA for passaging is widespread. Feeder-free culture depends on the use of specially formulated media and culture substrates that ensure adherence. These reagents entail an additional expense that may exceed some laboratory budgets. In addition, culture under feeder-free conditions has been associated with a perturbed differentiation potential due to the lack of specific factors in the feeder-free culture media and a resultant transition from the naive state to the primed state. Growth on mitotically arrested feeder cells avoids this transition and can bring overall costs down to a manageable level, thus facilitating the broader use of pluripotent stem cells in laboratory research.
Disclosures
Joel C. Glover is the director and Hege Brincker Fjerdingstad is the daily manager of the Norwegian Core Facility for Human Pluripotent Stem Cells. The authors have no competing financial interests or other conflicts of interest to disclose.
Acknowledgements
We thank Lars Moen for assistance during preliminary experiments and the Norwegian Core Facility for Human Pluripotent Stem Cells at the Norwegian Center for Stem Cell Research, Oslo University Hospital, for the use of facilities. The H9 hESC line was obtained from WiCell, and the HS429 hESC line was obtained from Outi Hovatta at the Karolinska Institute. Both were used in accordance with Material Transfer Agreements. The NCS001 and NCS002 hiPSC lines were generated by the Norwegian Core Facility for Human Pluripotent Stem Cells. That reprogramming and all the work reported here were performed with the approval of the Southeastern Norway Regional Ethics Committee (approval REK 2017/110).
Materials
| Name | Company | Catalog Number | Comments |
| 0.5 M EDTA pH 8.0 | Invitrogen | 15575020 | |
| 15 mL centrifuge tubes | Sarstedt | 62.554.502 | |
| 2-mercaptoethanol | Gibco | 31350-010 | |
| 50 mL centrifuge tubes | Sarstedt | 62.547.254 | |
| Basic fibroblast growth factor (bFGF) | PeproTech | AF-100-18B-250UG | |
| Brand Bürker Chamber | Fisher Scientific | 10628431 | |
| Disposable scalpels no.15 | Susann-Morton | 505 | |
| DPBS (1x) without Ca/Mg | Gibco | 14190-094 | |
| Easy Grip tissue culture dish, 35 x 10 mm | Falcon | 353001 | |
| Eppendorf pipette 1 mL | Eppendorf | ||
| Eppendorf pipette 200 μL | Eppendorf | ||
| FBS (Fetal Bovine Serum) | Gibco | 10270-106 | |
| Filter tip 1,000 μL | Sarstedt | 70.1186.210 | |
| Filter tip 200 μL | Sarstedt | 70.760.211 | |
| Gamma Cell 3000 ELAN irradiation machine (alternatively, use Mitomycin C to arrest proliferation) | Best Theratronics | BT/MTS 8007 GC3000E | |
| Glutamax 100x | Gibco | 35050-038 | |
| Growth Factor Reduced Matrixgel | Corning | 734-0269 | |
| H9 hESC line | WiCell | WAe009-A | |
| hPSC Genetic Analysis Kit | Stem Cell Technologies | #07550 | |
| HS429 hESC line | ECACC | KIe024-A | |
| Human Foreskin Fibroblasts -CRL2429 line | ATTC | CRL2429 | |
| IMDM (1x) | Gibco | 21980-032 | |
| iPSC lines | Norwegian Core Facility for Human Pluripotent Stem Cells | NCS001 & NCS002 | |
| Knockout DMEM | Gibco | 10829-018 | |
| Laser Scanning Confocal Microscope or equivalent (we use the LSM 700 from Zeiss) | Zeiss | ||
| Microscope | CETI | ||
| Mitomycin C | Sigma Aldrich | M4287 | |
| Non-essential amino acids (NEAA) | Gibco | 11140.035 | |
| Pipettes, plastic 10 mL | Sarstedt | 86.1254.001 | |
| Pipettes, plastic, 5 mL | Sarstedt | 86.1253.001 | |
| Serum Replacement (SR) | Gibco | 10828-028 | |
| Sterile filters 0.22 um | Sarstedt | 83.1826.102 | |
| T-75 culture flask | ThermoScientific | 156499 | |
| Trypan Blue Stain (0.4 %) | Gibco | 15250-061 | |
| Trypsin-EDTA, 500 mL | Gibco | 25300062 |
References
- Skottman, H., Hovet, O. Culture conditions for human embryonic stem cells. Reproduction. 132 (5), 691-698 (2006).
- Hovatta, O., et al. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Human Reproduction. 18 (7), 1404-1409 (2003).
- Desai, N., Rambhia, P., Gishto, A. Human embryonic stem cell cultivation: historical perspective and evolution of xeno-free culture systems. Reproductive Biology and Endocrinology. 13, 9(2015).
- Villa-Diaz, L. G., et al. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nature Biotechnology. 28 (6), 581-583 (2010).
- Watanabe, M., et al. TGFb superfamily signaling regulates the state of human stem cell pluripotency and capacity to create well-structured telencephalic organoids. Stem Cell Reports. 17 (10), 2220-2238 (2022).
- Garitaonandia, I., et al. Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PLoS One. 10 (2), e0118307(2015).
- Inzunza, J., et al. Derivation of human embryonic stem cell lines in serum replacement medium using postnatal human fibroblasts as feeder cells. Stem Cells. 23, 544-549 (2005).
- Watanabe, K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 25 (6), 681-686 (2007).
- Rivera, T., Zhao, Y., Ni, Y., Wang, J. Human-induced pluripotent stem cell culture methods under cGMP conditions. Current Protocols in Stem Cell Biology. 54 (1), 117(2020).
- Castro-Viñuelas, R., et al. Tips and tricks for successfully culturing and adapting human induced pluripotent stem cells. Molecular Therapy. Methods & Clinical Development. 23, 569-581 (2021).
- Meng, G., Rancourt, D. E. Derivation and maintenance of undifferentiated human embryonic stem cells. Methods in Molecular Biology. 873, 69-90 (2012).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved