A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Microfluidic Tools for Probing Fungal-Microbial Interactions at the Cellular Level
In This Article
Summary
Owing to the opacity of soil, interactions between its constituent microbes cannot easily be visualised with cellular resolution. Here, two microfluidic tools are presented, which offer new opportunities for investigating fungal-microbial interactions. The devices are versatile and simple to use, enabling high spatiotemporal control and high-resolution imaging at the cellular level.
Abstract
Filamentous fungi are successful inhabitants of soil and play a major role in soil ecosystems, such as in the decomposition of organic and inorganic matter, as well as regulation of nutrient levels. There they also find numerous opportunities to interact with a variety of other microbes such as bacteria or other fungi. Studying fungal interactions at the cellular level, however, can be challenging owing to the black box-like nature of soil. New microfluidic tools are being developed for the study of fungal interactions; two platforms designed to study bacterial-fungal and fungal-fungal interactions are highlighted. Within these microchannels, fungal-microbial interactions can be monitored in controlled physico-chemical environments at higher temporal and spatial resolution than previously possible. Application of these tools have yielded numerous novel biological insights, such as the observation of bacterial polar attachment to hyphae or revealing uncharacterised fungal-fungal antagonisms. A key feature of these methodologies regards the ease of use of this tool by non-experts, yielding highly translatable technologies for use in microbiology labs.
Introduction
Soil is an exceptionally diverse environment containing an abundance of microorganisms that are instrumental to carbon and phosphorous cycles1,2. Filamentous fungi are a major component of numerous ecosystems as decomposers of organic and inorganic matter and can enhance the nutrition of plants through the formation of symbiotic relationships3,4. Within soil, fungi interact dynamically with a multitude of microbes such as other fungi5, bacteria6, viruses7 and nematodes8. These interactions have significant consequences for soil and plant health. Yet, owing to a lack of appropriate experimental systems capable of imaging interacting microorganisms with high-resolution, many remain undefined.
Investigations concerning bacterial-fungal interactions (BFIs) and fungal-fungal interactions (FFIs) have valuable applications in a range of fields, including antimicrobials in medicine and biological control agents in agriculture. For example, the fungus Coprinopsis cinerea produces the peptide copsin, which has been shown to exhibit antibacterial activity against the human pathogen Listeria monocytogenes9. Similarly, the fungal-derived compound, griseofulvin, is widely used as a treatment for human fungal infections and is additionally able to inhibit the growth of the plant pathogenic fungus Alternaria solani10,11. Several strains of the soil-dwelling bacterium Bacillus subtilis have also been demonstrated to be effective biocontrol agents of the fungal plant pathogen Rhizoctonia solani12,13. Nonetheless, due to limitations associated with traditional methodologies, BFIs and FFIs are poorly understood at the level of single cells.
Conventional studies typically explore BFIs and FFIs on the macroscale using agar plates with two or more species in confrontation. Their interaction is assessed by measuring growth rates and metabolite production of the confronting species14,15,16; however, this methodology is only resolved to the colony level. To study interactions at the cellular level, bacterial and fungal inoculants can be cultivated on glass microscope slides coated with agar that are then imaged under a microscope17. Nevertheless, it can be difficult to follow a single hypha using microscope slides due to a lack of confinement, meaning time-lapse images are harder to obtain. Further, the opportunity to spatially confine other microorganisms within defined regions of the fungal mycelium or create defined chemical environments that can be perturbed, for example, is not possible in such set-ups. The "black box" nature of soil also adds to the complexity of studying fungal-microbial interactions at the level of single cells18. By observing interacting species away from the incredible diversity of the soil microbiome, the exact manner by which individual members interact can be assessed. Thus, there is a continued need for versatile platforms that enable high-resolution, single-cell imaging of BFIs and FFIs.
Microfluidic technologies, so-called lab-on-a-chip systems, provide an ideal platform for the study of BFIs and FFIs at the level of single cells. The field of microfluidics, originating from technologies developed for chemical analysis and microelectronics, has been adopted by the biological sciences19. Microfluidic technologies regulate small volumes of fluids within a bespoke network of miniaturised channels, having at least one dimension on the micrometre scale, and their use in biological research is expanding20. In particular, microfluidic devices have been developed to examine the growth of filamentous fungi21,22,23,24,25,26,27,28,29,30. One benefit of using this technology is that the confinement of hyphae and the distribution of nutrients within microchannels more closely resembles the structure of the soil environment than conventional agar methods31. Recently, microfluidic platforms have been used to investigate interactions between human neutrophils and fungal pathogens32, bacteria and plant roots33, as well as fungi and nematodes34,35.
One of the many advantages of using microfluidics for studying microbial interactions includes the specific control of the microchannel environment. For instance, laminar flow regimes can be exploited to generate defined concentration gradients, which is especially useful when examining bacterial chemotaxis36. Another advantage is that the transparent nature of poly(dimethylsiloxane) (PDMS), an inexpensive, biocompatible elastomeric polymer commonly used in the manufacture of microfluidic devices, facilitates high-resolution imaging of single cells using brightfield and fluorescence microscopy37. Likewise, the confinement of microbes within microchannels means that time-lapse experiments tracking single cells can be performed, allowing individual cellular responses to be recorded and quantified37. Lastly, as microfluidic devices can be designed to be user-friendly, they can be easily employed by non-experts38.
Furthering knowledge of the interactions between soil-dwelling microorganisms is important for improving sustainable ecosystem management practises that maintain biodiversity and to mitigate the impact of climate change on terrestrial environments39. Thus, the development of novel microfluidic tools is fundamental to expand understanding of fungi and their interactions at the cellular level. The protocol here will focus on two microfluidic devices produced for the study of BFIs40 and FFIs41 as represented in Figure 1.

Figure 1: Visual and schematic representation of the bacterial-fungal interaction (BFI) and fungal-fungal interaction (FFI) devices. (A) Image of the BFI device. A mycelial plug is placed at the entrance to one end of the microchannels to allow hyphal growth into the device. The bacterial inlet is at the opposite end. Scale bar = 5 mm. (B) Schematic overview of the BFI device, depicting the positioning of the bacterial inlets and direction of hyphal growth through the interaction microchannels. The channels are 10 µm in depth, 100 µm wide, and 7 mm long, with 28 observation channels in total. (C) Confrontation assay on agar plate between Coprinopsis cinerea and Bacillus subtilis NCIB 3610, scale bar = 20 mm (left). Microscopy images showing the interaction between C. cinerea and B. subtilis NCIB 3610 within the microchannel (middle and right), i.e., polar attachment of bacteria to fungal hyphae. Scale bar = 25 µm (middle) and 10 µm (right). (D) Image of the FFI device bonded to a glass-bottomed Petri dish, dual inoculated with mycelial plugs. Scale bar = 1 cm. (E) Schematic overview of the FFI device. Two fungal inoculant plugs are introduced into the inlets at either end of the device, permitting hyphal exploration of the microchannels. Control channels are connected to one fungal inlet only and have a dead-end channel, preventing interactions between the test fungi. Interaction channels connect both fungal inlets and permit hyphal interactions between the test subjects within the microchannel. Each interaction channel consists of 18 diamond-shaped sections, measuring a total length of 8.8 mm (490 x 430 µm per diamond), 10 µm deep, and having a connecting region between each diamond of 20 µm. Channel types are duplicated, scale bars = 1 mm. (F) Interaction zone between two approaching hyphal fronts, growing from opposite ends of the interconnected interaction channel. Phase contrast microscopy image, scale bar = 250 µm. The panels in this figure have been modified from Stanley et al., 2014 (A-C)40 and Gimeno et al., 2021 (D-F)41. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Protocol
NOTE: A summary of the procedures outlined in this protocol are visually depicted in Figure 2.
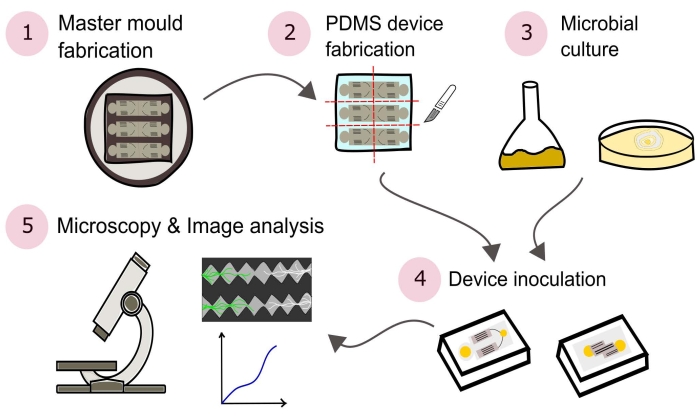
Figure 2: Schematic representation of the presented methodology consisting of five major sections detailed in this protocol. Device designs are created using computer aided design (CAD) software and a master mold manufactured using photolithography (1). This is used to cast poly(dimethylsiloxane) (PDMS), which is then diced into slabs and bonded to glass-bottomed Petri dishes to form the microfluidic devices (2). Microbes to be included in the study are cultured (3) and used to inoculate the devices (4). Interactions are studied using microscopy and quantified using image analysis techniques (5). Please click here to view a larger version of this figure.
1. Master mold fabrication
- Photomask production
- Generate microfluidic device designs using computer aided design (CAD) software. The dimensions of the presented devices are given in Figure 1 and more details about the specific design features are comprehensively listed in the respective publications40,41.
- Export the CAD design file using an appropriate format (e.g., .dwg, .dxf). Print a film photolithography mask by sending the exported CAD design file to a commercial provider for printing.
- Photolithography
NOTE: The following steps should be conducted within a dust-free and light-controlled environment, such as a laminar flow hood or clean room facility. The experimental conditions provided here are given as a guide and should be optimised in-house. The authors recommend seeking specific training and consulting established protocols42.- Prepare a 100 mm silicon wafer by baking in an oven at 200 °C for 2 h. Spin-coat the silicon wafer with SU-8 2010 photoresist, aiming for a target thickness of 10 µm, using the following conditions: 500 rpm for 10 s (acceleration 100 rpm/s) and 3,000 rpm for 45 s (acceleration 300 rpm/s).
CAUTION: SU-8 photoresist is hazardous, take care when handling and prevent inhalation and contact with skin. It is flammable, potentially carcinogenic and toxic to the environment. - Bake the coated silicon wafer at 95 °C for 2.5 min (soft bake). Expose the photoresist to ultraviolet (UV) light, using the film photolithography mask and an energy dose of 140 mJ/cm2 at 365 nm wavelength using a mask aligner.
- Bake the coated silicon wafer at 95 °C for 3.5 min (post exposure bake). Immerse and agitate the silicon wafer in developer solution for 3 min to reveal the microfabricated structures by removing the unexposed photoresist.
CAUTION: The developer solution can be flammable, take appropriate precautions when handling and storing. - Rinse with fresh developer solution for 10 s. Rinse with isopropyl alcohol for 10 s and air dry. Use filtered, compressed air to ensure structures are thoroughly dry. Measure the height of the SU-8 structures, for example, using a profilometer.
- Silanize each master mold with 50 µL of chlorotrimethylsilane by applying a vacuum pressure of 50 mbar for 2 h. The authors note that re-silanisation of the master mold was not found to be necessary.
CAUTION: Chlorotrimethylsilane is a hazardous substance. Wear appropriate personal protective equipment (PPE) and handle with care. Avoid contact with skin and eyes and prevent inhalation. Keep away from sources of ignition and use in a well-ventilated area.
- Prepare a 100 mm silicon wafer by baking in an oven at 200 °C for 2 h. Spin-coat the silicon wafer with SU-8 2010 photoresist, aiming for a target thickness of 10 µm, using the following conditions: 500 rpm for 10 s (acceleration 100 rpm/s) and 3,000 rpm for 45 s (acceleration 300 rpm/s).
2. Device fabrication
NOTE: The following steps should be conducted within a dust-free environment, such as a laminar flow hood.
- Preparation of poly(dimethylsiloxane) (PDMS) slabs
- Prepare approximately 40 g of PDMS by thoroughly mixing the base and curing agent in a 10:1 ratio using a spatula in a clean plastic cup. Degas the mixture to remove all air bubbles by placing the plastic cup containing the PDMS into a vacuum chamber (vacuum pressure = 50 mbar) for 1 h.
- Secure the master mold into a plastic mount using clear tape. Clean using compressed filtered air to remove any dust particles.
NOTE: Alternatively, aluminium foil can be shaped around a glass Petri dish, and then used to house the master mold and contain the PDMS43. - Pour the PDMS mixture onto the centre of the master mold, ensuring it is on a level surface, and allow to settle.
NOTE: The PDMS mixture should be poured as closely as possible to the surface of the master mold and a continuous flow maintained to minimize the introduction of air bubbles. Air bubbles can be removed by directing compressed air over the bubble or by scooping them out using a fine needle. - Cover the master mold loosely with a plastic lid to prevent dust particles from settling on the surface of the PDMS. Transfer the master mold to an oven and cure overnight at 70 °C.
- Remove the master mold from the oven and allow to cool. Peel the cured PDMS away from the master mold and plastic frame, taking care to avoid damaging the master mold/PDMS.
- Place clear tape over the microchannels embossed into the PDMS to maintain a dust free surface. Ensure the tape is removed prior to bonding.
- Cut the PDMS into slabs (i.e., if multiple devices are included in the design on the master mold, many can be fabricated from a single casting) as designated by the design using a mounted guillotine or razor blade. When cutting the lateral opening of the BFI PDMS slab, ensure that the microchannels are fully open (Figure 1A). For the FFI PDMS slab, ensure each corner is trimmed to enable it to fit into the glass-bottomed Petri dish shown in Figure 1D.
- Punch desired inlet/outlet holes according to the device design. Use a precision cutter to punch inlet holes of 3.18 mm and 4.75 mm for the exemplar BFI and FFI devices, respectively.
- Bonding of PDMS slabs to create devices
NOTE: The following washing steps (2.2.1-2.2.2) use an ultrasonic cleaner filled with purified water (ddH2O) at 37 kHz. Washing the PDMS slabs helps to enhance successful bonding44 and reduce the risk of contamination. To manipulate the PDMS slabs, use clean forceps and lift using the inlet holes to avoid damage to the microchannels or device surface.- Submerse the PDMS slabs in 0.5 M NaOH and sonicate for 5 min. Rinse with sterile ddH2O. Transfer PDMS slabs into 70% ethanol solution and sonicate for 5 min. Rinse with sterile ddH20.
- Immerse PDMS slabs into sterile ddH2O and sonicate for 5 min. Remove the PDMS slabs from the sterile ddH2O, dry using filtered compressed air, and place into a sterile square Petri dish.
- Place the square Petri dish containing the PDMS slabs into an oven at 70 °C for 1 h to dry. Remove from the oven and allow to cool in a dust-free environment. Clear any dust from the surface of the PDMS slabs using tape and/or filtered compressed air.
- Activate the surfaces of the PDMS slabs and glass-bottomed Petri dishes to be bonded using a plasma cleaner with the following settings: vacuum pressure 0.75 mbar, power 50%, coating time 1 min. Place the surfaces to be activated (and subsequently bonded) facing upward in the plasma cleaner.
- Remove the PDMS slabs and glass-bottomed Petri dishes from the plasma cleaner and bond by gently placing the activated surfaces into conformal contact with one another. Bond the BFI and FFI PDMS slabs to the 35 mm and 50 mm diameter glass-bottomed Petri dishes (glass thickness 0.17 mm), respectively.
NOTE: Take care not to apply too much pressure when bonding, as this can result in collapse of the microchannels. - Check for successful bonding by simply trying to pull the PDMS slab off the glass-bottomed Petri dish with tweezers. Visualize the devices by eye or using generic microscopy to ensure no collapse of the inoculation inlets or microchannels.
NOTE: For saturated conditions (i.e., water-saturated and/or nutrient-rich conditions), include step 2.2.7 of the protocol. If water-unsaturated conditions are required, proceed to step 2.2.8. Devices can be filled with water or media. - Fill the devices immediately after bonding by pipetting 100 µL of the desired solution for the BFI device (bacterial inlet and lateral opening) or 30 µL of media into each inlet (60 µL total) for the FFI device. If air bubbles are present, these will dissipate around 10 min after filling as PDMS is porous.
- Add sterile ddH2O (~100-200 µL) in the Petri dish to maintain humidity.
3. Microbial culture
NOTE: The following steps provide a general microbiological procedure for fungal and bacterial culture and should be carried out under sterile conditions (i.e., using a flame or microbiological safety cabinet) appropriate for the level of containment required for the desired microbes. Specific examples are given at the end of each section for a species of interest.
- Fungal culture
- Prepare desired culture medium supplemented with agar. Autoclave the medium at 121 °C for 15 min. Allow the medium to cool to 50 °C and pour into 9 cm diameter Petri dishes, maintaining sterile conditions.
- Use a cork borer to remove a 4 mm diameter plug of agar containing mycelium from a fridge stock colony of the desired fungal strain to activate the isolate. This is conducted to ensure standardized and vigorous growth of the fungus prior to device inoculation.
NOTE: The microbes can also be activated from a glycerol stock, i.e., fungal isolates stored on agar plugs in 50% glycerol solution at -70 °C41. - Place the side of the plug with mycelium in contact with the agar surface in the center of the uninoculated Petri dish. Replace the lid on top of the Petri dish and seal before incubating at the appropriate temperature for the desired strain for the required amount of time, typically, around 3 to 4 days.
NOTE: Example culture conditions for Trichoderma rossicum: Malt extract agar, incubated at 25 °C in the dark for 48 h.
- Bacterial culture
- Streak out the desired bacterial isolate from the source stock (e.g., glycerol stock or single colony from the agar plate) onto an agar plate to achieve single bacterial colonies and ensure no contamination45. Seal the plate with film.
- Incubate at a temperature and duration specific to the isolate of interest until individual colonies are observed.
- Prepare the desired culture broth. For instance, add 10 g of tryptone, 10 g of NaCl and 5 g of yeast extract per 1 L of ddH2O to prepare LB medium for the culture of B. subtilis. Autoclave the medium at 121 °C for 15 min.
- Allow the medium to cool to room temperature. Add the medium to a sterile culture flask inside a sterile environment.
- Touch a single bacterial colony from the agar plate using a sterile inoculation loop. Transfer the inoculated loop into the sterile culture medium by briefly touching the liquid with the loop.
- Seal the flask using a sterile lid or foil, and place inside a shaking incubator overnight using the settings appropriate for the selected species.
NOTE: Example culture conditions for B. subtilis: i) liquid culture - aerobic growth at 37 °C at 200 rpm in LB medium and ii) plate culture - room temperature on LB agar plate. Refer to the FFI/BFI papers40,41 for further details of culturing different fungal strains.
4. Device inoculation
NOTE: The following steps should take place inside a laminar flow hood using sterile equipment.
- Fungal inoculation
- Use a sterilized cork borer (ø = 4 mm) to remove an agar plug from the colony on the periphery of a 3-day-old culture (step 3.1). Ensure that the growing hyphal front remains intact.
- Introduce the plug into the fungal inlet, mycelium side down, with the growth direction of the hyphal front oriented toward the microchannel openings to encourage hyphal infiltration of the channels.
- Repeat steps 4.1.1-4.1.2 for the second fungal species (if using the FFI device), introducing the plug into the opposite inlet. If using the BFI device, omit this step and continue to step 4.1.4.
- Seal the Petri dish with transparent film and incubate at 25-28 °C in the dark until imaging commences. Determine the pre-imaging incubation time depending on the intended biological event to be observed, e.g., fungal-fungal confrontations, and the growth rate of included species within the device.
- Bacterial inoculation
- Dilute the bacteria from an overnight culture (step 3.2) in a 1:25 ratio using the same culture medium as detailed in step 3.2.3. Culture for 3 h at 37 °C.
- Wash the bacteria by pelleting culture using a centrifuge at 2000 x g for 10 min. Discard the supernatant and resuspend the cells in desired volume of 0.9% w/v sodium chloride solution.
- Centrifuge again to obtain a pellet. Discard the supernatant and resuspend the cells in liquid medium (e.g., C. cinerea minimal medium to an OD600 of 1). Optimize the OD600 value for the bacterial strain in question.
- Remove the BFI device from the incubator and open it in a sterile environment. Pipette 10 µL of suspension into the bacterial inlet.
NOTE: Optimise the exact inoculation timings for the bacterial-fungal interactions in question. For example, introduce bacteria into the BFI device 18 h post-fungal inoculation if using C. cinerea. - Seal the Petri dish with a transparent film and incubate at 25 °C in the dark until imaging commences. Store the device upright.
5. Microscopy and image analysis
- Microscopy
NOTE: The researcher should select the appropriate method of imaging concordant with the nature of the experiment to be conducted, for example, an inverted wide-field epifluorescence or confocal microscopy. A general overview has been provided here, as specific details will depend upon the attributes of the chosen microscopy set up.- Turn on the microscope computer, microscope main body (if applicable), camera, temperature-controlled incubator, and light source(s). Ensure that the microscope has been set up correctly, e.g., Köhler illumination has been correctly applied for even illumination of the sample. Initiate the imaging software package.
NOTE: When using a temperature-controlled incubator, it is important to allow the microscope temperature to equilibrate for several hours before starting an experiment. - Mount the microfluidic device into the stage insert. Ensure that the device is well secured, i.e., with tape, to prevent dislodging the device during active stage movement.
- Acquire images of the inoculated devices, e.g., either single-point or time-lapse experiments. Comprehensive imaging specifications relevant to the experiments conducted with the BFI and FFI devices are provided in the respective aforementioned publications40,41.
NOTE: Brightfield images were acquired by using phase contrast microscopy to visualize hyphal proliferation through the growth channels using autofocus software and either 10x magnification, 0.30 NA (numerical aperture) or 20x magnification, 0.45 NA objective lenses. Excitation of fluorescent reporter proteins was achieved using a high-power light emitting diode light engine with wavelengths specific to the fluorophore. - Export images into a suitable format for subsequent image processing. For example, .tiff.
- Turn on the microscope computer, microscope main body (if applicable), camera, temperature-controlled incubator, and light source(s). Ensure that the microscope has been set up correctly, e.g., Köhler illumination has been correctly applied for even illumination of the sample. Initiate the imaging software package.
- Image analysis
NOTE: The authors recommend Fiji46 as a tool for image analysis, but other software packages are available. The following are examples of image analyses conducted using Fiji from the presented BFI and FFI device publications. These steps are specific to a Mac and may differ slightly if using a PC.- Hyphal growth rate measurements
NOTE: This method was used in the BFI manuscript40 to measure the growth rates of individual hyphae.- Download, install, and launch Fiji. Import the image sequence from a time-lapse experiment by selecting File > Import > Image Sequence. Locate the folder where the data is stored and select Open. In the Sequence Options window, select Preferences, and then OK.
- Select the Straight Line icon from the main tool bar. Position the beginning of the straight line at the tip of the growing hyphal tip by clicking and then concomitantly dragging the cursor to another point within the window. A yellow line will appear with three boxes indicating the beginning, mid-point, and end of the line.
- Move to the next frame in the image sequence by pressing and holding Ctrl and >. Position the end of the straight line at the tip of the growing hypha by selecting and dragging the square box to the correct position.
- Press and hold Ctrl and M to measure the length of the line in pixels. A Results window will appear with the measured data. Define the data displayed in the Results window as follows: click on the Results window, and then select Results > Set Measurements.
- Move to the next frame in the image sequence by pressing and holding Ctrl and then >. Position the beginning of the straight line at the tip of the growing hypha by selecting and dragging the square box to the correct position.
- Press and hold Ctrl and then M to measure the length of the line in pixels. A Results window will appear with the measured data.
- Repeat steps 5.2.1.5-5.2.1.6 until finished measuring the growth of the hypha in pixels.
- Select all of the data in the Results window. Copy and paste into another software programme, e.g., a spreadsheet, to process the data. Plot the hyphal growth (in pixels or micrometres) as a function of time and calculate the average growth rates. Perform at least three biological replicates per experiment.
- Fluorescence intensity measurements
NOTE: This method was used in the FFI publication41 to assess the change in fluorescence intensity in hyphae of Fusarium graminearum 8/1-wt-GFP upon contact with Clonostachys rosea 016 as a function of time.- Download, install, and launch Fiji. Import image sequence from a time-lapse experiment by selecting File > Import > Image Sequence. Locate the folder where the data is stored and select Open. In the Sequence Options window select Preferences and then OK.
- Specify a region of interest (ROI) to measure the absolute fluorescence intensity of a hypha using the rectangular tool, located in the main tool bar. The size of the square can be defined exactly as follows: Edit > Selection > Specify; the ROI can also be saved for future reference in the ROI Manager by selecting Edit > Selection > Add to Manager.
- Measure the absolute fluorescence intensity (mean gray value) within the defined ROI for each image in the entire image sequence or stack as follows: Image > Stacks > Measure Stack. The Results window will open automatically once all of the images in the stack have been processed.
NOTE: The data displayed in the Results window can be defined as follows: click on the Results window and then select Results > Set Measurements. Ensure that Mean Gray Value has been selected. - Select all of the data in the Results window. Copy and paste into another software programme, e.g., a spreadsheet, to plot the absolute fluorescence intensities of the specified ROI as a function of time.
- Repeat steps 5.2.2.2-5.2.2.4 to collect absolute fluorescence intensity measurements for each ROI, i.e., on the hypha of interest, next to the hypha of interest or within the corresponding control channel.
- Calculate the appropriate relative fluorescence intensities in arbitrary units (AU), e.g., by dividing the absolute fluorescence intensity of the ROI [hypha of interest] by the absolute fluorescence intensity of the ROI [control channel]. Consult the FFI publication41 for more specific details.
- Perform at least three biological replicates per experiment and plot the relative fluorescence intensities as a function of time.
- Hyphal growth rate measurements
Access restricted. Please log in or start a trial to view this content.
Results
Representative results are presented from the exemplar BFI40 and FFI41 devices. Hyphal growth rate measurements can easily be obtained using these devices in combination with basic microscopy techniques. Figure 3A-B illustrates bacterial-fungal interactions between C. cinerea hyphae and B. subtilis NCIB 3610. The presence of B. subtilis halts the growth of C. cinerea after ca...
Access restricted. Please log in or start a trial to view this content.
Discussion
This article presents a protocol for the study of fungal-microbial interactions using channel microfluidics. The authors aim to demonstrate the versatility of these devices and encourage adaptation to suit the researcher's interests. Using the exemplar BFI and FFI devices, fungal-microbial interactions can be studied in more detail than previously accessible. By removing the background complexity and heterogeneity of the soil, moderating the growth of hyphae such that they are confined to a single monolayer, and tigh...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare no competing interests.
Acknowledgements
We acknowledge financial support from the Department of Bioengineering at Imperial College London and The Leverhulme Trust (Research Grant Reference: RPG-2020-352).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Agar | Difco Laboratories | 214010 | Used to solidify culture medium for bacterial and fungal cultivation within Petri dishes |
| Aluminum foil | Fisher Scientific Ltd | 11759408 | |
| AutoCAD 2021 | Autodesk, USA | ||
| Autoclave (VX-75) | Systec | ||
| Centrifuge (5810R) | Eppendorf | ||
| Chlorotrimethysilane | Merck Life Sciences | 386529 | CAUTION: Chlorotrimethylsilane is a hazardous substance. Wear appropriate PPE and handle with care. Avoid contact with skin and eyes and prevent inhalation. Keep away from sources of ignition and use in a well-ventilated area. |
| Cork borer | SLS | COR1000 | |
| Developer solution (mr-Dev 600) | Microresist Technologies | CAUTION: mr-Dev 600 developer solution is flammable | |
| Erlenmeyer flasks | VWR | 214-1108 | e.g. 200 mL; choose size to suit your exact needs |
| Ethanol (70% v/v) | Fisher Scientific Ltd | E/0650DF/15 | Diluted from 99.8% (Analytical Reagent Grade) |
| Fiji | ImageJ | Exemplar software package for imaging processing | |
| Filtered, compressed air | Available as standard in most labs. Altervatively, an oil-free compressor with air regulator can be used. | ||
| Flat-headed wafer tweezers | SLS | INS5026 | |
| Forceps | Fisher Scientific Ltd | 10008051 | Bent, sharp |
| Glass bottom petri dish | World Precision Instruments | FD35-100 | 35 mm |
| Glass bottom petri dish | World Precision Instruments | FD5040-100 | 50 mm |
| Glass crystallisation dishes | VWR | 216-1865 | Used for washing of PDMS slabs |
| Glass crystallisation dishes | VWR | 216-1866 | Used in the development of master moulds |
| Glass media bottles | Fisher Scientific Ltd | 15456113 | e.g. 250 mL; choose size to suit your exact needs |
| Glass syringe (Hamilton) | Fisher Scientific Ltd | 10625251 | Used for dispensing chlorotrimethylsilane |
| Hot plate (HP 160 III BM) | SAWATEC | ||
| Inoculation loop | VWR | COPA175CS01 | |
| Isopropyl alcohol | Sigma-Aldrich | W292907 | |
| Laminar flow hood | Air Science (PCR) | Exemplar laminar flow hood used for device fabrication | |
| LB medium | Fisher Scientific Ltd | BP9723-500 | Exemplar nutrient broth for bacterial overnight culture |
| Light emitting diode light engine (LedHUB) | Omicron-Laserage Laserprodukte GmbH | Exemplar light source that can be used for imaging fungal-microbial interactions (fluorescence) | |
| MA6 Ultraviolet mask aligner | Suss Microtec | ||
| Malt extract | VWR | 84618 | Used to make exemplar fungal culture medium (Malt extract agar) |
| Mask Writer | Applied Materials | 4700DP | Example of a mask writer which can be used to print photo-mask for photolithography |
| Master mould plastic mount | 3D-printed bespoke holder manufactured in-house | ||
| Microbiological safety cabinet (BioMat2) | Contained Air Solutions | Exemplar MSC used for microbial culture and device inoculation | |
| Milli-Q purified water | Available as standard in biology labs. | ||
| NaOH | Fisher Scientific Ltd | BP359-500 | |
| NIS-Elements Advanced Research imaging software | Nikon | Exemplar software package for image acquisition | |
| NIS-Elements Free Viewer | Nikon | Exemplar software package for viewing acquired images | |
| Oven (Binder BD115) | Fisher Scientific Ltd | 15602126 | Used for curing poly(dimethylsiloxane)(PDMS) |
| Oven (CLO-2AH-S) | KOYO | Used for preparing silicon wafers | |
| Parafilm | Bemis | HS234526B | transparent film |
| Petri dishes, square sterile | Fisher Scientific Ltd | 11708573 | 120.5 mm |
| Petri dishes, sterile | Fisher Scientific Ltd | 15370366 | 90 mm |
| Photolithography mask | Micro Lithography Services Ltd. UK | ||
| Plasma cleaner (Zepto) | Diener Electronic | 100012601 | |
| Plastic cup | Semadeni | 8323 | |
| Plastic spatula | Semadeni | 3340 | |
| Portable precision balance (OHAUS Scout) | Fisher Scientific Ltd | 15519631 | Used for weighing PDMS, media components etc. |
| Precision cutter | Syneo | HS1251135P1183 | Cutting edge diameter: 3.18 mm |
| Precision cutter | Syneo | HS1871730P1183S | Cutting edge diameter: 4.75 mm |
| Profilometer | Bruker | Dektak XT-stylus | |
| Razor blades | Häberle Labortechnik | 9156110 | |
| Refridgerator | Haden | 4-6 °C | |
| Retiga R1 CCD camera | Qimaging | Exemplar camera that can be used for imaging fungal-microbial interactions | |
| Scotch magic tape | Office Depot | 3969954 | 19 mm invisible tape; clear tape |
| Shaking incubator (Cole-Parmer SI500) | Fisher Scientific Ltd | 10257954 | |
| Silicon wafer | Inseto | 100 mm | |
| Soda lime glass plate | Inseto | 125 mm x 125 mm x 2 mm. Used to hold photolithography mask in mask aligner | |
| Sodium chloride | Sigma-Aldrich | S7653 | |
| Spincoater | SAWATEC | SM-180-BM | |
| SU-8 2010 photoresist | MicroChem | CAUTION: SU-8 photoresist is hazardous, take care when handling and prevent inhalation and contact with skin. Flammable, potentially carcinogenic and toxic to the environment. | |
| Sylgard 184 elastomer kit | VWR | 634165S | Used for the preparation of poly(dimethylsiloxane)(PDMS) devices |
| Temperature controlled incubator | Okolab | Exemplar incubator that can be used for imaging fungal-microbial interactions | |
| Ti2-E inverted epifluorescence microscope | Nikon | MEA54000 | Exemplar microscope that can be used for imaging fungal-microbial interactions |
| Ultrasonic cleaner S-Line | Fisher Scientific Ltd | FB15050 | |
| Vacuum desiccator | Fisher Scientific Ltd | 10528861 | Silianisation and PDMS degassing should be conducted in separate desiccators |
| x10/0.3 NA CFI Plan Fluor DL objective lens | Nikon | MRH20105 | Exemplar objective lens that can be used for imaging fungal-microbial interactions |
| x20/0.45 NA CFI Plan Fluor DL objective lens | Nikon | MRH48230 | Exemplar objective lens that can be used for imaging fungal-microbial interactions |
References
- Zhu, Y. -G., Miller, R. M. Carbon cycling by arbuscular mycorrhizal fungi in soil-plant systems. Trends in Plant Science. 8 (9), 407-409 (2003).
- Dai, Z., et al. Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. The ISME Journal. 14 (3), 757-770 (2020).
- Op De Beeck, M., et al. Regulation of fungal decomposition at single-cell level. The ISME Journal. 14 (4), 896-905 (2020).
- Bender, S. F., et al. Symbiotic relationships between soil fungi and plants reduce N2O emissions from soil. The ISME Journal. 8 (6), 1336-1345 (2014).
- Dullah, S., et al. Melanin production and laccase mediated oxidative stress alleviation during fungal-fungal interaction among basidiomycete fungi. IMA Fungus. 12 (1), 33(2021).
- Deveau, A., et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiology Reviews. 42 (3), 335-352 (2018).
- Bian, R., et al. Facilitative and synergistic interactions between fungal and plant viruses. Proceedings of the National Academy of Sciences of the United States of America. 117 (7), 3779-3788 (2020).
- Jiang, X., Xiang, M., Liu, X. Nematode-trapping fungi. Microbiology Spectrum. 5 (1), (2017).
- Essig, A., et al. a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. Journal of Biological Chemistry. 289 (50), 34953-34964 (2014).
- Tang, H. -Y., Zhang, Q., Li, H., Gao, J. -M. Antimicrobial and allelopathic metabolites produced by Penicillium brasilianum. Natural Product Research. 29 (4), 345-348 (2015).
- Bai, Y. -B., et al. Antifungal activity of griseofulvin derivatives against phytopathogenic fungi In vitro and In vivo and three-dimensional quantitative structure-activity relationship analysis. Journal of Agricultural and Food Chemistry. 67 (22), 6125-6132 (2019).
- Solanki, M. K., et al. Characterization of antagonistic-potential of two Bacillus strains and their biocontrol activity against Rhizoctonia solani in tomato. Journal of Basic Microbiology. 55 (1), 82-90 (2015).
- Jamali, H., Sharma, A., Srivastava, A. K. Biocontrol potential of Bacillus subtilis RH5 against sheath blight of rice caused by Rhizoctonia solani. Journal of Basic Microbiology. 60 (3), 268-280 (2020).
- Válková, H., Novotný, Č, Malachová, K., Šlosarčíková, P., Fojtík, J. Effect of bacteria on the degradation ability of Pleurotus ostreatus. Science of The Total Environment. 584-585, 1114-1120 (2017).
- Leyva-Rojas, J. A., Coy-Barrera, E., Hampp, R. Interaction with soil bacteria affects the growth and amino acid content of Piriformospora indica. Molecules. 25 (3), Basel, Switzerland. 572(2020).
- Dullah, S., et al. Fungal interactions induce changes in hyphal morphology and enzyme production. Mycology. 12 (4), 279-295 (2021).
- Marfetán, J. A., Romero, A. I., Folgarait, P. J. Pathogenic interaction between Escovopsis weberi and Leucoagaricus sp.: mechanisms involved and virulence levels. Fungal Ecology. 17, 52-61 (2015).
- Cortois, R., De Deyn, G. B. The curse of the black box. Plant and Soil. 350 (1), 27-33 (2012).
- Whitesides, G. M. The origins and the future of microfluidics. Nature. 442 (7101), 368-373 (2006).
- Sackmann, E. K., Fulton, A. L., Beebe, D. J. The present and future role of microfluidics in biomedical research. Nature. 507 (7491), 181-189 (2014).
- Hanson, K. L., et al. Fungi use efficient algorithms for the exploration of microfluidic networks. Small. 2 (10), 1212-1220 (2006).
- Held, M., Edwards, C., Nicolau, D. V. Probing the growth dynamics of Neurospora crassa with microfluidic structures. Fungal Biology. 115 (6), 493-505 (2011).
- Thomson, D. D., et al. Contact-induced apical asymmetry drives the thigmotropic responses of Candida albicans hyphae. Cellular Microbiology. 17 (3), 342-354 (2015).
- Lee, K. K., Labiscsak, L., Ahn, C. H., Hong, C. I. Spiral-based microfluidic device for long-term time course imaging of Neurospora crassa with single nucleus resolution. Fungal Genetics and Biology. 94, 11-14 (2016).
- Asenova, E., Lin, H. Y., Fu, E., Nicolau, D. V., Nicolau, D. V. Optimal fungal space searching algorithms. IEEE Transactions on NanoBioscience. 15 (7), 613-618 (2016).
- Soufan, R., et al. Pore-scale monitoring of the effect of microarchitecture on fungal growth in a two-dimensional soil-like micromodel. Frontiers in Environmental Science. 6, (2018).
- Uehling, J. K., et al. Microfluidics and metabolomics reveal symbiotic bacterial-fungal interactions between Mortierella elongata and Burkholderia include metabolite exchange. Frontiers in Microbiology. 10, 2163(2019).
- Millet, L. J., et al. Increasing access to microfluidics for studying fungi and other branched biological structures. Fungal Biology and Biotechnology. 6 (8), 1-14 (2019).
- Baranger, C., Fayeulle, A., Le Goff, A. Microfluidic monitoring of the growth of individual hyphae in confined environments. Royal Society Open Science. 7 (8), 191535(2020).
- Aleklett, K., Ohlsson, P., Bengtsson, M., Hammer, E. C. Fungal foraging behaviour and hyphal space exploration in micro-structured Soil Chips. The ISME Journal. 15 (6), 1782-1793 (2021).
- Aleklett, K., et al. Build your own soil: exploring microfluidics to create microbial habitat structures. The ISME Journal. 12 (2), 312-319 (2018).
- Ellett, F., Jorgensen, J., Frydman, G. H., Jones, C. N., Irimia, D. Neutrophil interactions stimulate evasive hyphal branching by Aspergillus fumigatus. PLOS Pathogens. 13 (1), 1006154(2017).
- Massalha, H., Korenblum, E., Malitsky, S., Shapiro, O. H., Aharoni, A. Live imaging of root-bacteria interactions in a microfluidics setup. Proceedings of the National Academy of Sciences of the United States of America. 114 (17), 4549-4554 (2017).
- Schmieder, S. S., et al. Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Current Biology. 29 (2), 217-228 (2019).
- Tayyrov, A., Stanley, C. E., Azevedo, S., Künzler, M. Combining microfluidics and RNA-sequencing to assess the inducible defensome of a mushroom against nematodes. BMC Genomics. 20 (1), 243(2019).
- Stanley, C. E., Grossmann, G., Casadevall i Solvas, X., deMello, A. J. Soil-on-a-Chip: microfluidic platforms for environmental organismal studies. Lab on a Chip. 16 (2), 228-241 (2016).
- Stanley, C. E., vander Heijden, M. G. A. Microbiome-on-a-Chip: new frontiers in plant-microbiota research. Trends in Microbiology. 25 (8), 610-613 (2017).
- Ortseifen, V., Viefhues, M., Wobbe, L., Grünberger, A. Microfluidics for biotechnology: bridging gaps to foster microfluidic applications. Frontiers in Bioengineering & Biotechnology. 8, 589074(2020).
- Jansson, J. K., Hofmockel, K. S. The soil microbiome-from metagenomics to metaphenomics. Current Opinion in Microbiology. 43, 162-168 (2018).
- Stanley, C. E., et al. Probing bacterial-fungal interactions at the single cell level. Integrative Biology (Camb). 6 (10), 935-945 (2014).
- Gimeno, A., et al. A versatile microfluidic platform measures hyphal interactions between Fusarium graminearum and Clonostachys rosea in real-time. Communications Biology. 4 (1), 262(2021).
- Duffy, D. C., McDonald, J. C., Schueller, O. J. A., Whitesides, G. M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Analytical Chemistry. 70 (23), 4974-4984 (1998).
- Stanley, C. E., et al. Fabrication and use of the dual-flow-RootChip for the imaging of Arabidopsis roots in asymmetric microenvironments. Bio-protocol. 8 (18), 3010(2018).
- Choi, C. -H., Lee, H., Weitz, D. A. Rapid patterning of PDMS microfluidic device wettability using syringe-vacuum-induced segmented flow in nonplanar geometry. ACS Applied Materials & Interfaces. 10 (4), 3170-3174 (2018).
- Sanders, E. R. Aseptic laboratory techniques: plating methods. Journal of Visualized Experiments. (63), e3064(2012).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Harting, R., et al. Pseudomonas strains induce transcriptional and morphological changes and reduce root colonization of Verticillium spp. Frontiers in Microbiology. 12, 652468(2021).
- Boenisch, M. J. Structural and molecular characterisation of the penetration process of Fusarium graminearum during Fusarium head blight infection. , Staats-und Universitätsbibliothek Hamburg Carl von Ossietzky. (2013).
- Eynck, C., Koopmann, B., Grunewaldt-Stoecker, G., Karlovsky, P., von Tiedemann, A. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. European Journal of Plant Pathology. 118 (3), 259-274 (2007).
- Ghanem, N., Stanley, C. E., Harms, H., Chatzinotas, A., Wick, L. Y. Mycelial effects on phage retention during transport in a microfluidic platform. Environmental Science & Technology. 53 (20), 11755-11763 (2019).
- Alrifaiy, A., Lindahl, O. A., Ramser, K. Polymer-based microfluidic devices for pharmacy, biology and tissue engineering. Polymers. 4 (3), 1349-1398 (2012).
- Duncombe, T. A., Tentori, A. M., Herr, A. E. Microfluidics: reframing biological enquiry. Nature Reviews Molecular Cell Biology. 16 (9), 554-567 (2015).
- Hoelzle, D., et al. Microfluidic device design, fabrication, and testing protocols. Protocol Exchange. , (2015).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved