A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Realistic Membrane Modeling Using Complex Lipid Mixtures in Simulation Studies
In This Article
Summary
Membrane lipid diversity in structure and composition is an important contributor to cellular processes and can be a marker of disease. Molecular dynamics simulations allow us to study membranes and their interactions with biomolecules at atomistic resolution. Here, we provide a protocol to build, run, and analyze complex membrane systems.
Abstract
Lipids are structural building blocks of cell membranes; lipid species vary across cell organelles and across organisms. This variety results in different mechanical and structural properties in the membrane that directly impact the molecules and processes that occur at this interface. Lipid composition is dynamic and can serve to modulate cell signaling processes. Computational approaches are increasingly used to predict interactions between biomolecules and provide molecular insights to experimental observables. Molecular dynamics (MD) is a technique based on statistical mechanics that predicts the movement of atoms based on the forces that act on them. MD simulations can be used to characterize the interaction of biomolecules. Here, we briefly introduce the technique, outline practical steps for beginners who are interested in simulating lipid bilayers, demonstrate the protocol with beginner-friendly software, and discuss alternatives, challenges, and important considerations of the process. Particularly, we emphasize the relevance of using complex lipid mixtures to model a cell membrane of interest to capture the appropriate hydrophobic and mechanical environments in simulation. We also discuss some examples where membrane composition and properties modulate the interactions of bilayers with other biomolecules.
Introduction
Lipids are major constituents of membranes, which provide boundaries for cells and enable intracellular compartmentalization1,2,3. Lipids are amphiphilic, with a polar head group and two hydrophobic fatty acid tails; these self-assemble into a bilayer to minimize contact of the hydrophobic chains with water3,4. Various combinations of hydrophilic head groups and hydrophobic tails result in different classes of lipids in biological membranes, such as glycerophospholipids, sphingolipids, and sterols (Figure 1)1,5,6. Glycerophospholipids are primary building blocks of eukaryotic cell membranes composed of glycerophosphate, long-chain fatty acids, and head groups of low-molecular weight7. Lipid nomenclature is based on differences in head groups; examples include phosphatidyl-choline (PC), phosphatidyl-ethanolamine (PE), phosphatidyl-serine (PS), phosphatidyl-glycerol (PG), phosphatidyl-inositol (PI), or the unmodified phosphatidic acid (PA)5,6. As for hydrophobic tails, the length and degree of saturation vary, along with the backbone structure. The possible combinations are numerous, resulting in thousands of lipid species in mammalian cells6. Changes in membrane lipid composition lead to different mechanical and structural membrane properties that impact the activity of both integral membrane proteins and peripheral proteins2,6.
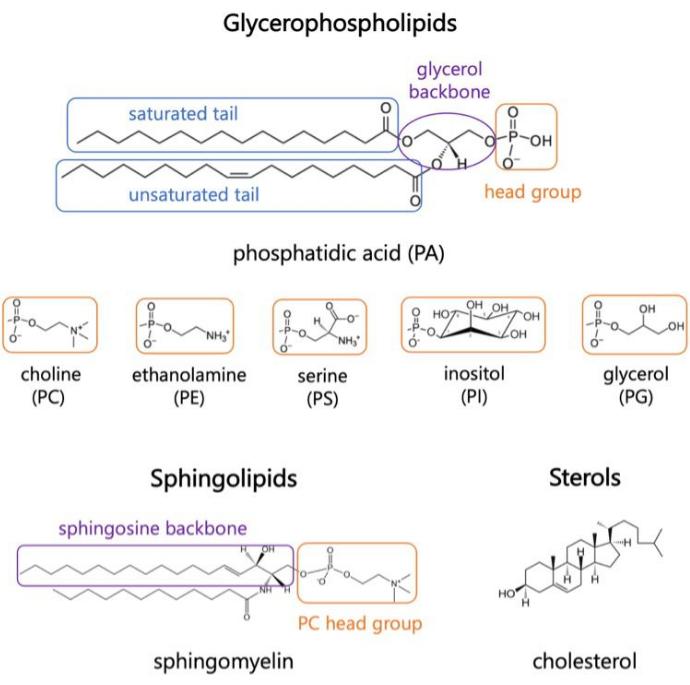
Figure 1. Representative lipid structures. Fatty acid tails are shown in blue boxes, common lipid head groups in orange, and sample backbones in purple. Please click here to view a larger version of this figure.
Lipids are active players in cellular processes, protein activation in signaling cascades, and healthy cell homeostasis8,9. Altered lipid dynamics are the result of infection or can be markers of pathogenesis of disease10,11,12,13,14,15. As barriers for the cell, the study of membrane lipids and their role in permeation of small molecules is of relevance for drug delivery systems and membrane disruption mechanisms16,17. Chemical diversity and different ratios of lipid species across organelles, tissues, and organisms give rise to complex membrane dynamics2. It is therefore important to retain these characteristics in modelling studies of lipid bilayers, especially when the goal of a study is to examine interactions of other biomolecules with the membrane. The lipid species to consider in a model depends on the organism and cellular compartment of interest. For example, PG lipids are important for electron transfer in photosynthetic bateria18, while phosphorylated inositol lipids (PIPs) are major players in plasma membrane (PM) dynamics and signaling cascades in mammalian cells19,20. Inside the cell, the PM, endoplasmic reticulum (ER), Golgi, and mitochondrial membranes contain unique lipid abundances that influence their function. For example, the ER is the hub for lipid biogenesis and transports cholesterol out to the PM and Golgi; it contains a high lipid diversity with an abundance of PC and PE, but low sterol content, which promotes membrane fluidity21,22,23,24. In contrast, the PM incorporates hundreds and even thousands of lipid species depending on the organism25, it contains high levels of sphingolipids and cholesterol that give it a characteristic rigidity compared to other membranes in the cell24. Leaflet asymmetry should be considered for membranes like the PM, which has an outer leaflet rich in sphingomyelin, PC, and cholesterol, and an inner leaflet rich in PE, PI and PS that are important for signaling cascades24. Finally, lipid diversity also prompts the formation of micro-domains that differ in packing and internal order, known as lipid rafts24,26; these exhibit lateral asymmetry, are hypothesized to play important roles in cellular signaling26, and are hard to study due to their transient nature.
Experimental techniques such as fluoroscopy, spectroscopy, and model membrane systems like giant unilamellar vesicles (GUVs) have been used to investigate interactions of biomolecules with membranes. However, the complex and dynamic nature of the components involved is difficult to capture with experimental methods alone. For example, there are limitations on the imaging of transmembrane domains of proteins, the complexity of membranes used in such studies, and the identification of intermediate or transient states during the process of interest27,28,29. Since the advent of molecular simulation of lipid monolayers and bilayers in the 1980s29, lipid-protein systems and their interactions can now be quantified at the molecular level. Molecular dynamics (MD) simulation is a common computational technique that predicts the movement of particles based on their intermolecular forces. An additive interaction potential describes the bonded and non-bonded interactions between particles of the system30. The set of parameters used to model these interactions is termed the simulation forcefield (FF). These parameters are obtained from ab initio computations, semi-empirical, and quantum mechanical calculations, and optimized to reproduced data from X-ray and electron diffraction experiments, NMR, infrared, Raman and neutron spectroscopy, amongst other methods31.
MD simulations can be used to study systems at various levels of resolution32,33,34. Systems that aim to characterize specific biomolecular interactions, hydrogen bonds, and other high-resolution details are studied with all-atom (AA) simulations. In contrast, coarse-grained (CG) simulations lump atoms into larger functional groups to reduce computational cost and examine larger scale dynamics33. Situated in between these two are united-atom (UA) simulations, where hydrogen atoms are combined with their respective heavy atoms to accelerate the computation33,35. MD simulations are a powerful tool for exploration of the dynamics of lipid membranes and their interactions with other molecules and can serve to provide molecular level mechanisms for processes of interest at the membrane interface. Additionally, MD simulations can serve to narrow down experimental targets and predict macromolecular properties of a given system based on microscopic interactions.
In brief, given a set of initial coordinates, velocities, and a set of conditions like constant temperature and pressure, positions and velocities of each particle are calculated through numerical integration of the interaction potential and Newton's Law of motion. This is repeated iteratively, thereby generating a simulation trajectory30. These calculations are performed with an MD engine; among several open-source packages, GROMACS36 is one of the most commonly used engines and the one we describe here. It also includes tools for analysis and constructing initial coordinates of systems to be simulated37. Other MD engines include NAMD38; CHARMM39, and AMBER40, which the user can select at their own discretion based on computational performance of a given system. It is critical to visualize the trajectories during the simulation as well as for analysis and interpretation of the results. A variety of tools are available; here we discuss visual molecular dynamics (VMD) that offers a wide range of features, including three dimensional (3-D) visualization with expansive drawing and coloring methods, volumetric data visualization, building, preparing, and analyzing trajectories of MD simulation systems, and trajectory-movie making with no limits on system size, if the memory is available41,42,43.
The accuracy of predicted dynamics between system components is directly influenced by the FF chosen for the propagation of the trajectory. Empirical FF parametrization efforts are pursued by few research groups. The most established and common FF for MD include CHARMM39, AMBER40, Martini44, OPLS45, and SIRAH46. The all-atom additive CHARMM36 (C36) force field47 is widely used for AA MD of membrane systems as it accurately reproduces experimental structural data. It was originally developed by the CHARMM community, and it is compatible with multiple MD engines like GROMACS and NAMD. Despite improvements across common FFs, there is a continuous effort to improve the parameter sets to allow predictions that closely reproduce experimental observables, driven by interests in particular systems of study48,49.
A challenge when simulating lipid membranes is determining the length of simulation trajectory. This is largely dependent on the metrics to be analyzed and the process that one aims to characterize. Typically, complex lipid mixtures require longer time to reach equilibrium, as more species must have enough time to diffuse on the membrane plane and reach a stable lateral organization. A simulation is said to be at equilibrium when the property of interest has reached a plateau and fluctuates about a constant value. It is common practice to obtain at least 100-200 ns of equilibrated trajectory to carry out appropriate statistical analysis on the properties and interactions of interest. It is common to run membrane-only simulations between 200-500 ns, depending on the complexity of the lipid mixture and research question. Protein-lipid interactions typically require longer simulation times, between 500-2000 ns. Some approaches to accelerate sampling and observable dynamics with membrane systems are: (i) the highly mobile membrane mimetic (HMMM) model, which replaces end carbons of lipids in the membrane with organic solvent to accelerate sampling50; and (ii) hydrogen mass repartitioning (HMR), which combines a fraction of the masses of heavy atoms within a system with those of hydrogen atoms to allow the use of a larger simulation timestep51.
The following protocol discusses a beginner-friendly approach to build, run, and analyze realistic membrane models using AA MD. Given the nature of MD simulations, multiple trajectories must be run to account for reproducibility and proper statistical analysis of the results. It is current practice to run at minimum three replicas per system of interest. Once the lipid species have been selected for the organism and process of interest, basic steps to build, run, and analyze a simulation trajectory of a membrane-only system are outlined and summarized in Figure 2.
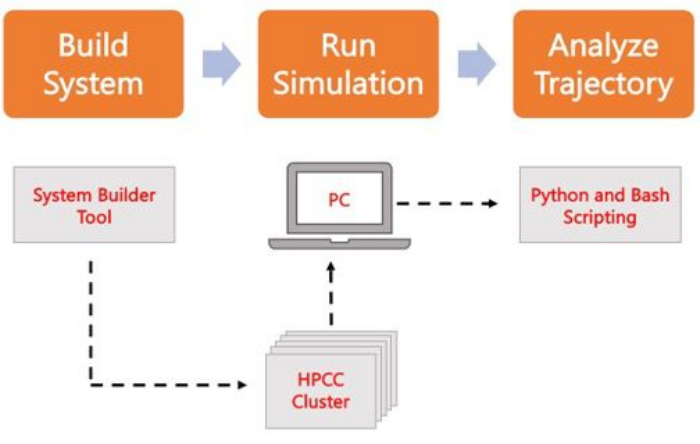
Figure 2. Schematic to run MD simulations. Orange boxes correspond to the three main steps described in the protocol. Underneath is the workflow of the simulation process. During system setup, the system containing the initial coordinates of a solvated membrane system is built with a system input generator like CHARMM-GUI Membrane Builder. After transferring the input files to a high-performance computing cluster, the simulation trajectory is propagated using an MD engine, such as GROMACS. Trajectory analysis can be done on the computer cluster or a local working station along with visualization. Analysis is then carried out, using either packages with built-in analysis code such as GROMACS and VMD, or by using Bash scripts or various Python libraries. Please click here to view a larger version of this figure.
Protocol
1. Building the system coordinates
- Navigate to CHARMM-GUI.org (C-GUI) using a web browser. On the top menu, navigate to Input Generator, then select Membrane Builder from the vertical options on the left side of the screen.
- To build a bilayer, select Bilayer Builder.
NOTE: First time users must activate their free account before building their first set of coordinates. - Select Membrane Only System. Save the generated JOB ID to retrieve the system and continue from where you left off during the process if needed.
- Visualize the systems during every step of the building process by clicking View Structure in the box located at the top of the page, or by downloading the resulting PDB file. Look out for missing components, mistakes in the selected input lipid species or patch size.
- Select the components of the system.
- Choose the Heterogeneous Lipid option, even if building a single-component bilayer; then select a Rectangular Box type.
- Select 45 water molecules per lipid for the hydration option; this is sufficient to ensure a fully hydrated bilayer.
- Set the length of XY to be based on number of lipid components. Then, select the number of lipids to be included for each lipid species determined ahead of the model. For the case study discussed in the next section, a membrane model with 600 lipids distributed symmetrically in two leaflets was built. To model the ER of eukaryotic cells a mixture of 336 DOPC, 132 DPPE, 60 CHOL, 72 POPI lipids was used for the PI model; and 330 DOPC, 126 DPPE, 54 CHOL, 66 POPI, and 24 DOPS lipids for the PI-PS model.
NOTE: C-GUI provides a library of lipid structures to choose from; click on the images next to the species name for its chemical structure. - Type the desired number of molecules in the upper and lower leaflet in the two boxes next to the lipid name. For the case study, a symmetric membrane composition is desired - make sure there are no errors about unmatched number of lipids in the upper leaflet and lower leaflet. If asymmetry is desired, make sure the total number of lipids in each leaflet is correct. For details on building asymmetric bilayers, refer to work by Park et al.52,53.
- Go to the top of the lipid species list and click on Show the System Info button. Assemble components and complete the system.
- Select the option to include neutralizing ions using the distance-based algorithm for faster convergence54.
- Leave the default solution concentration of KCl at 0.15 mM. This is a typical salt concentration to render neutral the simulation box for membrane bilayers.
NOTE: If a different concentration is to be used, make sure to click the Calculate Solvent Composition button after editing it.
- Select the simulation conditions and settings.
- Select CHARMM36m as the FF option; it is commonly used for lipid and protein simulations, but the user may select other options discussed in the introduction.
- Select GROMACS as the MD engine to obtain sample input files in the corresponding format.
NOTE: GROMACS is recommended for new users because it has multiple online resources, tutorials, and forums for support. The user can select from multiple MD engines to explore options in terms of simulation performance and code syntax. - Select the Constant Particle-Pressure-Temperature (NPT) ensemble, by far the most used dynamic ensemble in the simulation of lipid bilayers.
- Set the temperature and pressure in Kelvin and bars as 303 K and 1 bar, respectively. It is typical to set the temperature between 298 K and 310 K for the study of biological processes to ensure a bilayer in the liquid disorder state.
NOTE: The temperature depends on the conditions of the process to be simulated and can be altered as needed. Depending on the lipid species in the model, set the temperature to be above the transition temperature of pure lipid components prior to running the simulation.
- Download the resulting files and transfer to the computer cluster.
- Visualize the final system on a software of choice, such as VMD or PyMol, and inspect for proper setup.
NOTE: It is good to check, for example, that there is sufficient water around the membrane such that lipids do not interact with image atoms during the simulation, and proper leaflet setup (a bilayer with no space or water in-between).
- Visualize the final system on a software of choice, such as VMD or PyMol, and inspect for proper setup.
2. Running MD simulations
- Upload and unzip the files from C-GUI on your computing cluster. Navigate to the Gromacs directory. Create a relaxation submission script.
- Follow the guidelines of the cluster for the format of a submission script.
- Copy the commands listed until just above the # Production comment in the README file into the submission script.
NOTE: This default from C-GUI is a loop that runs a 6-step relaxation of the system. If a different and well-established protocol is desired, edit it to read the coordinates just built and downloaded from C-GUI.
- Copy the commands listed until just above the # Production comment in the README file into the submission script.
- Submit the relaxation script and confirm that all output files have been downloaded for all steps before moving to the production run. Upon completion, check for the following output files from GROMACS, generated during the 6 step run: *.log, *.tpr, *.gro, *.edr, *.trr / *.xtc
- Create a production run script.
- Use one of the samples gmx grompp and gmx mdrun commands from any of the relaxation steps as a template.
- Before using the script, make sure to create a *.mdp file that contains similar simulation options as the provided step7_production.mdp file.
NOTE: The provided default options are standard for membrane simulations; ditances are listed in nm and time is given in picoseconds or number of steps (picoseconds / integration time step). Update the nsteps to run up to the desired simulation length (this is equal to dt * nsteps) and nst[x,v,f]out to update the data saving frequency in number of integration steps. For the case study, set the nsteps to 250,000,000 for a simulation length of 500ns (simulation time / integration step = 500,000 ps / 0.002ps), and nst[x,v,f]out to 50,000 to save data every 100 ps
- Before running the actual simulation, run benchmark studies to determine the best use of resources.
- Run the system for 1-2 ns using different number of computing nodes.
NOTE: The ER case study was submitted on the UB center for computational research (CCR) high-performance computing cluster55 for 2 ns, where the performance was tested for 1-10 nodes. - Compare the performance in ns/day for each setting to determine the optimal resources for the run. It is common practice to select the number of nodes that result in 75%-80% of maximum performance.
- Run the system for 1-2 ns using different number of computing nodes.
- Run the production run.
- Run each system in triplicates to ensure reproducibility and conduct statistical analysis on the data.
- Extend the trajectory based on the benchmarks if the allowed queue time for submission on the computing cluster runs out. Use the gmx convert-tpr, then gmx mdrun commands to continue trajectory collection.
NOTE: Options are described in the GROMACS documentation online (https://manual.gromacs.org/). - For a membrane-only system, examine if the system has reached equilibrium by calculating the area-per-lipid over time. If it has not, extend the simulation trajectory.
3. Analyzing the trajectory
- Visualize the system before running analysis to determine molecules of interest and portion of the trajectory that is intended for characterization.
- Compress raw trajectory files (*.trr) by changing file format to *.xtc and/or skipping frames to reduce file size and facilitate more efficient transfer to the local station for visualization and analysis.
NOTE: For large membrane systems, one may opt to strip water from the trajectory to further reduce the file size. This can be done with index files on GROMACS, TCL scripts on VMD, or Python libraries such as MDAnalysis and MDTraj. - Perform chosen analyses during the equilibrated portion of the trajectory, as determined from the area-per-lipid timeseries.
NOTE: Refer to discussion for more details on typical membrane analyses and how to run them.
Results
To illustrate the use of the protocol and the results that can be obtained, a comparison study for membrane models for the endoplasmic reticulum (ER) is discussed. The two models in this study were (i) the PI model, which contains the top four lipid species found in the ER, and (ii) the PI-PS model, which added the anionic phosphatidylserine (PS) lipid species. These models were later used in a study of a viral protein and how it interacts with the membrane, the interest on PS has been cited as important for the per...
Discussion
Experimental techniques can visualize biomolecules at high resolution using cryo-electron microscopy (cryo-EM)58, fluorescence techniques, and atomic force microscopy (AFM)59. However, it is challenging to capture the interplay and dynamics of molecular interactions that underlie biological pathways, disease pathogenesis, and therapeutic delivery at the atomic or amino acid level. Here, capabilities of MD simulations to study lipid membranes and the main steps to design, bu...
Disclosures
The authors have no competing interests to disclose.
Acknowledgements
The authors thank Jinhui Li and Ricardo X. Ramirez for their simulation trajectories and discussions during the writing of this manuscript. O.C. was supported by the University at Buffalo Presidential Fellowship and National Institute of Health's Initiative for Maximizing Student Development Training Grant 1T32GM144920-01 awarded to Margarita L. Dubocovich (PI).
Materials
| Name | Company | Catalog Number | Comments |
| Anaconda3 | Anaconda Inc (Python & related libraries) | N/A | |
| CHARMM-GUI.org | Im lab, Lehigh University | N/A | |
| GROMACS | GROMACS development team | N/A | |
| Linux HPC Cluster | UB CCR | N/A | |
| MATLAB | MathWorks | N/A | |
| VMD | Theoretical and Computational Biophysics Group | N/A |
References
- Vanni, S., Riccardi, L., Palermo, G., De Vivo, M. Structure and Dynamics of the Acyl Chains in the Membrane Trafficking and Enzymatic Processing of Lipids. Accounts of Chemical Research. 52 (11), 3087-3096 (2019).
- Harayama, T., Riezman, H. Understanding the diversity of membrane lipid composition. Nature Reviews Molecular Cell Biology. 19 (5), 281-296 (2018).
- Tanaka, M., Edward, H. .. . E. g. e. l. m. a. n. . Comprehensive Biophysics. , 261-272 (2012).
- Bruce Alberts, A. J., Julian Lewis, , Martin Raff, , Keith Roberts, , Peter Walter, . Molecular Biology of the Cell. , (2002).
- Watson, H. Biological membranes. Essays in Biochemistry. 59, 43-69 (2015).
- Coskun, &. #. 2. 2. 0. ;., Simons, K. Cell Membranes: The Lipid Perspective. Structure. 19 (11), 1543-1548 (2011).
- Douglas G, H. a. y. e. s., Daniel, K. Y., Solaiman, D., Richard, . Biobased Surfactants (Second Edition) eds. , 515-529 (2019).
- González-Rubio, P., Gautier, R., Etchebest, C., Fuchs, P. F. J. Amphipathic-Lipid-Packing-Sensor interactions with lipids assessed by atomistic molecular dynamics. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1808, 2119-2127 (2011).
- Halbleib, K., et al. Activation of the Unfolded Protein Response by Lipid Bilayer Stress. Molecular Cell. 67, 673-684 (2017).
- Andreasen, M., Lorenzen, N., Otzen, D. Interactions between misfolded protein oligomers and membranes: A central topic in neurodegenerative diseases. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1848 (9), 1897-1907 (2015).
- Calianese, D. C., Birge, R. B. Biology of phosphatidylserine (PS): basic physiology and implications in immunology, infectious disease, and cancer. Cell Commununication and Signaling. 18 (1), 41 (2020).
- Nieto-Garai, J. A., Contreras, F. X., Arboleya, A., Lorizate, M. Role of Protein-Lipid Interactions in Viral Entry. Advanced Biology. 6, 2101264 (2022).
- Mazzon, M., Mercer, J. Lipid interactions during virus entry and infection. Cell Microbiology. 16, 1493-1502 (2014).
- Colombelli, C., Aoun, M., Tiranti, V. Defective lipid metabolism in neurodegeneration with brain iron accumulation (NBIA) syndromes: not only a matter of iron. Journal of Inherited Metabolic Disease. 38 (1), 123-136 (2015).
- Saini-Chohan, H. K., Mitchell, R. W., Vaz, F. M., Zelinski, T., Hatch, G. M. Delineating the role of alterations in lipid metabolism to the pathogenesis of inherited skeletal and cardiac muscle disorders: Thematic Review Series: Genetics of Human Lipid Diseases. Journal of Lipid Research. 53 (1), 4-27 (2012).
- Martinotti, C., Ruiz-Perez, L., Deplazes, E., Mancera, R. L. Molecular Dynamics Simulation of Small Molecules Interacting with Biological Membranes. ChemPhysChem. 21 (14), 1486-1514 (2020).
- Li, J., Kalyanram, P., Rozati, S., Monje-Galvan, V., Gupta, A. Interaction of Cyanine-D112 with Binary Lipid Mixtures: Molecular Dynamics Simulation and Differential Scanning Calorimetry Study. ACS Omega. 7 (11), 9765-9774 (2022).
- Nagy, L., et al. Protein/Lipid Interaction in the Bacterial Photosynthetic Reaction Center: Phosphatidylcholine and Phosphatidylglycerol Modify the Free Energy Levels of the Quinones. Biochemistry. 43 (40), 12913-12923 (2004).
- Ramirez, R. X., Campbell, O., Pradhan, A. J., Atilla-Gokcumen, G. E., Monje-Galvan, V. Modeling the molecular fingerprint of protein-lipid interactions of MLKL on complex bilayers. Frontiers in Chemistry. 10, (2023).
- Dondelinger, Y., et al. MLKL Compromises Plasma Membrane Integrity by Binding to Phosphatidylinositol Phosphates. Cell Reports. 7 (4), 971-981 (2014).
- van Meer, G., Voelker, D. R., Feigenson, G. W. Membrane lipids: where they are and how they behave. Nature Reviews Molecular Cell Biology. 9 (2), 112-124 (2008).
- van Meer, G., de Kroon, A. I. P. M. Lipid map of the mammalian cell. Journal of Cell Science. 124 (1), 5 (2011).
- Lee, H. R., Lee, G. Y., You, D. G., Kim, H. K., Young, D. Y. Hepatitis C virus p7 induces membrane permeabilization by interacting with phosphatidylserine. International Journal of Molecular Sciences. 21 (3), 897 (2020).
- Casares, D., Escribá, P. V., Rosselló, C. A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. International Journal of Molecular Sciences. 20 (9), 2167 (2019).
- Marrink, S. J., et al. Computational Modeling of Realistic Cell Membranes. Chemical Reviews. 119 (9), 6184-6226 (2019).
- Janmey, P. A., Kinnunen, P. K. J. Biophysical properties of lipids and dynamic membranes. Trends in Cell Biology. 16 (10), 538-546 (2006).
- Brémaud, E., Favard, C., Muriaux, D. Deciphering the Assembly of Enveloped Viruses Using Model Lipid Membranes. Membranes. 12, 441 (2022).
- Campbell, O., Monje-Galvan, V. Protein-driven membrane remodeling: Molecular perspectives from Flaviviridae infections. Biophysical Journal. 122 (11), 1890-1899 (2022).
- Loschwitz, J., Olubiyi, O. O., Hub, J. S., Strodel, B., Poojari, C. S. Computer simulations of protein-membrane systems. Progress in molecular biology and translational science. 170, 273-403 (2020).
- Shell, M. S., Scott Shell, M. . Thermodynamics and Statistical Mechanics: An Integrated ApproachCambridge Series in Chemical Engineering. , 21-49 (2015).
- Yang, J., et al. Molecular Dynamic Simulation of Ni-Al Alloy-H2O Reactions Using the ReaxFF Reactive Force Field. ACS Omega. 8 (11), 9807-9814 (2023).
- Ingólfsson, H. I., Arnarez, C., Periole, X., Marrink, S. J. Computational 'microscopy' of cellular membranes. Journal of Cell Science. 129 (2), 257-268 (2016).
- Klauda, J. B. Perspective: Computational modeling of accurate cellular membranes with molecular resolution. The Journal of Chemical Physics. 149 (22), 220901 (2018).
- Chavent, M., Duncan, A. L., Sansom, M. S. P. Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Current Opinion in Structural Biology. 40, 8-16 (2016).
- Khakbaz, P., Monje-Galvan, V., Zhuang, X., Klauda, J. B., Otto Geiger, . Biogenesis of Fatty Acids, Lipids and Membranes. , 1-19 (2017).
- Abraham, M. J., et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1, 19-25 (2015).
- Lemkul, J. A. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package. Living Journal of Computational Molecular Science. 1 (1), 5068 (2018).
- Phillips, J. C., et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. The Journal of Chemical Physics. 153 (4), 044130 (2020).
- Klauda, J. B., et al. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. The Journal of Physical Chemistry B. 114 (23), 7830-7843 (2010).
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., Case, D. A. Development and testing of a general amber force field. Journal of Computational Chemistry. 25 (9), 1157-1174 (2004).
- John Stone, A. A., et al. . Using VMD. , (2011).
- Humphrey, W., Dalke, A., Schulten, K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 14 (1), 33-38 (1996).
- Hsin, J., Arkhipov, A., Yin, Y., Stone, J. E., Schulten, K. Using VMD: An Introductory Tutorial. Current Protocols in Bioinformatics. 24 (1), 5.7.1-5.7.48 (2008).
- Souza, P. C. T., et al. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nature Methods. 18 (4), 382-388 (2021).
- Jorgensen, W. L., Maxwell, D. S., Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. Journal of the American Chemical Society. 118 (45), 11225-11236 (1996).
- Machado, M. R., et al. The SIRAH 2.0 Force Field: Altius, Fortius, Citius. Journal of Chemical Theory and Computation. 15 (4), 2719-2733 (2019).
- Huang, J., et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nature Methods. 14 (1), 71-73 (2017).
- Mu, J., Liu, H., Zhang, J., Luo, R., Chen, H. F. Recent Force Field Strategies for Intrinsically Disordered Proteins. Journal of Chemical Information and Modeling. 61 (3), 1037-1047 (2021).
- Inakollu, V. S. S., Geerke, D. P., Rowley, C. N., Yu, H. Polarisable force fields: what do they add in biomolecular simulations. Current Opinion in Structural Biology. 61, 182-190 (2020).
- Ohkubo, Y. Z., et al. Accelerating Membrane Insertion of Peripheral Proteins with a Novel Membrane Mimetic Model. Biophysical Journal. 102 (9), 2130-2139 (2012).
- Hopkins, C. W., Le Grand, S., Walker, R. C., Roitberg, A. E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. Journal of Chemical Theory and Computation. 11 (4), 1864-1874 (2015).
- Park, S., Beaven, A. H., Klauda, J. B., Im, W. How Tolerant are Membrane Simulations with Mismatch in Area per Lipid between Leaflets. Journal of Chemical Theory and Computation. 11 (7), 3466-3477 (2015).
- Park, S., Im, W., Pastor, R. W. Developing initial conditions for simulations of asymmetric membranes: a practical recommendation. Biophysical Journal. 120 (22), 5041-5059 (2021).
- Wu, E. L., et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. Journal of Computational Chemistry. 35 (27), 1997-2004 (2014).
- Center for Computational Research, U.a.B.. . CCR Facility Description. , (2019).
- Piggot, T. J., Allison, J. R., Sessions, R. B., Essex, J. W. On the Calculation of Acyl Chain Order Parameters from Lipid Simulations. Journal of Chemical Theory and Computation. 13 (11), 5683-5696 (2017).
- Li, J., Monje-Galvan, V. Effect of Glycone Diversity on the Interaction of Triterpenoid Saponins and Lipid Bilayers. ACS Applied Bio Materials. , (2023).
- Renaud, J. P., et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nature Reviews Drug Discovery. 17 (7), 471-492 (2018).
- Ando, T., Uchihashi, T., Kodera, N. High-Speed AFM and Applications to Biomolecular Systems. Annual Review of Biophysics. 42 (1), 393-414 (2013).
- Martínez, L., Andrade, R., Birgin, E. G., Martínez, J. M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. Journal of Computational Chemistry. 30 (13), 2157-2164 (2009).
- Jewett, A. I., et al. Moltemplate: A Tool for Coarse-Grained Modeling of Complex Biological Matter and Soft Condensed Matter Physics. Journal of Molecular Biology. 433 (11), 166841 (2021).
- Jo, S., Kim, T., Iyer, V. G., Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. Journal of Computational Chemistry. 29 (11), 1859-1865 (2008).
- Polêto, M. D., Lemkul, J. A. Integration of experimental data and use of automated fitting methods in developing protein force fields. Communications Chemistry. 5 (1), 38 (2022).
- Hynninen, A. P., Crowley, M. F. New faster CHARMM molecular dynamics engine. Journal of Computational Chemistry. 35 (5), 406-413 (2014).
- Kim, S. Issues on the Choice of a Proper Time Step in Molecular Dynamics. Physics Procedia. 53, 60-62 (2014).
- Grubmüller, H., Heller, H., Windemuth, A., Schulten, K. Generalized Verlet Algorithm for Efficient Molecular Dynamics Simulations with Long-range Interactions. Molecular Simulation. 6 (1-3), 121-142 (1991).
- Darden, T., York, D., Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. Journal of Chemical Physics. 98 (12), 10089-10092 (1993).
- . . Hepatitis C. , (2021).
- Braun, E., et al. Best Practices for Foundations in Molecular Simulations [Article v1.0]. Living Journal of Computational Molecular Science. 1 (1), 5957 (2018).
- Moradi, S., Nowroozi, A., Shahlaei, M. Shedding light on the structural properties of lipid bilayers using molecular dynamics simulation: a review study. RSC Advances. 9 (8), 4644-4658 (2019).
- Monje-Galvan, V., Klauda, J. B. Modeling Yeast Organelle Membranes and How Lipid Diversity Influences Bilayer Properties. Biochemistry. 54 (45), 6852-6861 (2015).
- Michaud-Agrawal, N., Denning, E. J., Woolf, T. B., Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. Journal of Computational Chemistry. 32 (10), 2319-2327 (2011).
- Gowers, R., et al. MDAnalysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations. SciPy. , (2016).
- McGibbon, R. o. b. e. r. t. . T., et al. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophysical Journal. 109 (8), 1528-1532 (2015).
- Fortunato, M. E., Colina, C. M. pysimm: A python package for simulation of molecular systems. SoftwareX. 6, 7-12 (2017).
- Scherer, M. K., et al. PyEMMA 2: A Software Package for Estimation, Validation, and Analysis of Markov Models. Journal of Chemical Theory and Computation. 11 (11), 5525-5542 (2015).
- Song, W., et al. PyLipID: A Python Package for Analysis of Protein-Lipid Interactions from Molecular Dynamics Simulations. Journal of Chemical Theory and Computation. 18 (2), 1188-1201 (2022).
- Monje-Galvan, V., Klauda, J. B. Peripheral membrane proteins: Tying the knot between experiment and computation. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1858 (7, Part B), 1584-1593 (2016).
- Monje-Galvan, V., Voth, G. A. Binding mechanism of the matrix domain of HIV-1 gag on lipid membranes. eLife. 9, e58621 (2020).
- Wang, B., Guo, C. Concentration-Dependent Effects of Cholesterol on the Dimerization of Amyloid-β Peptides in Lipid Bilayers. ACS Chemical Neuroscience. 13 (18), 2709-2718 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved