Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
GC-based Detection of Aldononitrile Acetate Derivatized Glucosamine and Muramic Acid for Microbial Residue Determination in Soil
W tym Artykule
Podsumowanie
We describe a method protocol for the GC-based analysis of the aldonitrile acetate derivatives of glucosamine and muramic acid extracted from soil. For elucidation of the chemical mechanism, we also present a strategy to confirm the structure of the derivative and the ion fragments formed upon electron ionization.
Streszczenie
Quantitative approaches to characterizing microorganisms are crucial for a broader understanding of the microbial status and function within ecosystems. Current strategies for microbial analysis include both traditional laboratory culture-dependent techniques and those based on direct extraction and determination of certain biomarkers1, 2. Few among the diversity of microbial species inhabiting soil can be cultured, so culture-dependent methods introduce significant biases, a limitation absent in biomarker analysis.
The glucosamine, mannosamine, galactosamine and muramic acid have been well served as measures of both the living and dead microbial mass, of these the glucosamine (most abundant) and muramic acid (uniquely from bacterial cell) are most important constituents in the soil systems3, 4. However, the lack of knowledge on the analysis restricts the wide popularization among scientific peers. Among all existing analytical methods, derivatization to aldononitrile acetates followed by GC-based analysis has emerged as a good option with respect to optimally balancing precision, sensitivity, simplicity, good chromatographic separation, and stability upon sample storage5.
Here, we present a detailed protocol for a reliable and relatively simple analysis of glucosamine and muramic acid from soil after their conversion to aldononitrile acetates. The protocol mainly comprises four steps: acid digestion, sample purification, derivatization and GC determination. The step-by-step procedure is modified according to former publications6, 7. In addition, we present a strategy to structurally validate the molecular ion of the derivative and its ion fragments formed upon electron ionization. We applied GC-EI-MS-SIM, LC-ESI-TOF-MS and isotopically labeled reagents to determine the molecular weight of aldononitrile acetate derivatized glucosamine and muramic acid; we used the mass shift of isotope-labeled derivatives in the ion spectrum to investigate ion fragments of each derivatives8. In addition to the theoretical elucidation, the validation of molecular ion of the derivative and its ion fragments will be useful to researchers using δ13C or ion fragments of these biomarkers in biogeochemical studies9, 10.
Protokół
1. Sample Preparation and Acid Extraction
- Freeze-dry soil samples after field collection.
- Grind and homogenize soil samples using a ball mill, soil grinder, or a mortar and pestle.
- Weigh soil samples (containing > 0.3 mg N) into a 25 mL hydrolysis flask.
- Add 10 mL 6M HCl into each hydrolysis flask, fill N2 gas in the flasks, and cap tightly.
- Hydrolyze at 105 °C in an incubator for 8 hours using an auto timer switch.
2. Sample Purification
- Remove the flasks from the incubator; cool to room temperature.
- Add 100 μL internal standard myo-inositol (1 mg mL-1 in water) to each flask; mix by swirling.
- Set up plastic funnels draining into 200 mL pear shaped flasks with ST/NS 24/40 joints, set on plastic cups for stability.
- Fold Whatman #2 Qualitative Circles (11 cm diameter) into quarters and set into funnels.
- Swirl each hydrolysis flask, and pour slurry into funnel to filter (you may further rinse each flask with ~3 mL of water).
- Dry the filtrate using a rotary evaporator at ~45 °C, applying vacuum.
- Resuspend the dried residue from each pear flask with 3~5 mL water (use ultrasonic bath if desired), and pour into a 40 mL Teflon tube; rinse the flask with a second aliquot of water.
- Adjust the pH to 6.6 - 6.8 using 1M KOH solution to precipitate metal ions and other organic molecules.
- Remove the precipitates by centrifugation at 2000 rcf for 10 minutes.
- Pour the supernatant into a 40 mL glass tube, cover the tube opening with parafilm, freeze at -20 °C, then poke holes in the parafilm.
- Freeze-dry the frozen supernatant to remove all liquid.
- Dissolve the residue with 3 mL dry methanol, vortexing thoroughly (use ultrasonic bath if desired); cap the tube, and then centrifuge at 2000 rcf for 10 minutes to settle out salts.
- Transfer the supernatant to a 3 mL conical reaction vial; evaporate to dryness by RapidVap machine at 45 °C (or under a gentle stream of dry nitrogen gas if desired).
- To each vial, add 100 μL recovery standard N-methylglucamine (1 mg mL-1 in water) and 1 mL H2O, cover the vial mouths with parafilm, freeze at -20 °C, perforate the parafilm, and then freeze dry.
- Make standards: add 100 μL muramic acid (0.5 mg mL-1 in methanol), 100 μL glucosamine (1 mg mL-1 in water), 100 μL myo-inositol (1 mg mL-1 in water), 100 μL N-methylglucamine (1 mg mL-1 in water), and 1 mL H2O, parafilm each vial, freeze at -20 °C, perforate the parafilm, then freeze-dry.
3. Derivatization
- Prepare derivatization reagent containing 32 mg mL-1 hydroxylamine hydrochloride and 40 mg mL-1 4-dimethylamino-pyridine in pyridine-methanol (4:1 v/v).
- Add 300 μL of the derivatization reagent to each of the reactivials, cap tightly, and vortex thoroughly.
- Put the vials in 75-80 °C water bath for 35 minutes (well sealed the vials to avoid allowing any water to enter the vials).
- Remove vials from the water bath, and cool to room temperature.
- Add 1 mL acetic anhydride to each of the 3 mL reactivials, cap tightly, and vortex thoroughly, then reheat in 75-80 °C water bath for 25 minutes.
- Remove vials from the water bath, and cool to room temperature.
4. Separation and Measurement
- Add 1.5 mL dichloromethane to each vial, cap tightly, and vortex thoroughly.
- Add 1 mL 1M HCl to each vial, cap tightly and vortex thoroughly to allow the solutions to sit undisturbed until the two phrases separate, aspirate and discard the top (aqueous) phase using 1000 μL pipettor.
- In the same fashion, extract the organic phase 3 times but with 1 mL H2O (with the last washing step, take special care to ensure that all of the aqueous top phase has been removed).
- Dry the final solution using RapidVap at 45 °C (or dry using nitrogen gas if desired).
- Dissolve in 300 μL ethyl acetate-hexane (1:1 v/v) and transfer to 2 mL amber screw-cap vials with a small-volume insert and cap tightly.
- For quantification, analyze by GC-FID using a fused silica nonpolar capillary column: 30 m long, 0.25 mm i.d., 0.25 um film thickness; stationary phase 5% phenyl-, 95% methyl-polysiloxane (DB-5 or equivalent) with hydrogen or helium as the carrier gas, at 0.5 mL min-1 constant flow. The chromatography is better on premium "inert" phases and hydrogen carrier gas, but is acceptable on the more common varieties and with helium. The detector settings are 300 °C; 400 mL min-1 air and 30 mL min-1 for both the nitrogen and hydrogen makeup gases. The injection and oven parameters are as follows: 1 μL split injection (30:1) with the GC inlet set at 250 °C; initial oven temperature, 120 °C; hold 1 min.; increase the oven temperature at 10 °C min-1 to 250 °C; hold 2.5 min.; ramp to 270 °C at 20 °C min-1; hold 2 minutes; ramp to 290 °C at 20 °C min-1, hold 5 min. Adjust the carrier gas flow rate so that the inositol, glucosamine and muramic acid derivatives elute at 250 °C, and the N-methylglucamine elutes at 270 °C.
5. Derivative Validation
- Use soft ionization LC-ESI-TOF-MS to identify the molecular ion and determine the molecular weight of the derivative.
- Use GC-EI-MS-SIM or GC-EI-MSMS for sensitivity enhancement to investigate targeted ions of the derivative.
- Use multiple isotopically labeled reagents for preparation of the derivatives, and then use the mass shift of those isotopically labeled derivatives in MS to investigate molecular ion and ion fragments.
6. Representative Results
The protocol of the method mainly comprises four steps: acid digestion, sample purification, derivatization and GC determination (Figure 1). An example of the analysis for glucosamine and muramic acid from standard stocks and from a soil sample is shown in Figure 2. Besides glucosamine and muramic acid, mannosamine and galactosamine (two isomers of glucosamine) can also be determined simultaneously using the method. Based on response factors of standards with respect to the internal standard myo-inositol, we can quantify these biomarkers in soil samples. The recovery standard has been used to qualitatively monitor the derivatization process. The schemes for formation of the aldononitrile acetate derivatized glucosamine and muramic acid are shown in Figure 3.
The proposed structures of the derivatives were determined by GC-EIMS-SIM for sensitivity enhancement, or soft ionization LC-ESI-TOF-MS8; the proposed structures of the ion fragments formed upon electron ionization were studied by comparing the ion spectra of the samples prepared with various isotope incorporations11. The Figure 4 shows the mass shift of the dominant ion m/z 187 of aldononitrile acetate derivatized glucosamine in three scenarios, prepared with non-labeled agents, D-acetic anhydrite, and U-13C-glucosamine. Other detailed information and explanations can be referred to our recent publications8, 11. This strategy could serve as a model to study derivative chemistry.
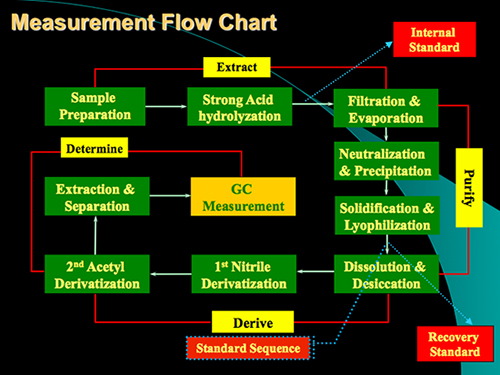
Figure 1. Measurement flow chart of the protocol for analysis of glucosamine and muramic acid in soil samples. The protocol mainly contains 4 steps: Acid digestion, Purification, Derivatization and GC determination.

Figure 2. GC-FID chromatograms of aldononitrile acetates of inositol, glucosamine, muramic acid, mannosamine, galactosamine and methylglucamine for standards and soil.
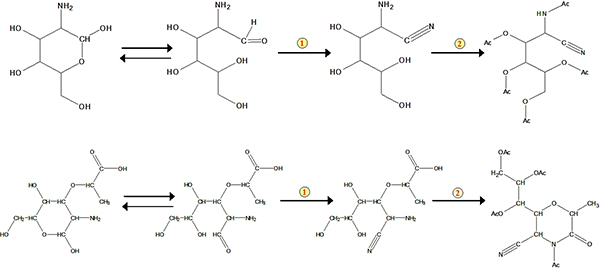
Figure 3. Schemes for formation of the aldononitrile acetate derivatized glucosamine and muramic acid. The number 1 represents nitrile reaction. The number 2 represents acetylation.

Figure 4. Mass spectra of aldononitrile acetate derivatized glucosamine associated with the dominant ion structures under EI mode prepared with (A) non-labeled agents, (B) D-acetic anhydrite, (C) U-13C-glucosamine. Star represents heavy isotope atom or isotope group. Please click here to see a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Dyskusje
The presented GC-based method for the analysis of aldononitrile acetate derivatized glucosamine and muramic acid provides a relatively rapid method to quantify these amino sugars, extracted from soil. The derivatives are chemically stable, and can be determined in one analysis. The method is not restricted to soil samples, and can be simplified for samples from non-soil matrix.
The vacuum pump used in this method is built to be resistant to acid. We further suggest setting up a base trap to p...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
We have nothing to disclose.
Podziękowania
This work was supported by grants from DOE Great Lakes Bioenergy Research Center (DOE BER office of Science DE-FC02-07ER64494). We are grateful to Dr. Xudong Zhang and his group members for helpful technical discussions and invaluable comments on finalizing the protocol.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Muramic acid | Sigma-Aldrich | M2503-25MG | |
| D-(+)-glucosamine hydrochloride | Sigma-Aldrich | G1514-100G | |
| N-methyl-D-glucamine | Sigma-Aldrich | M2004-500G | |
| Myo-inositol | Fisher Scientific | A307003G025 | |
| Methanol (dry) | Acros Organics | AC326950010 | |
| 4-dimethylamino-pyridine | Acros Organics | AC148270050 | |
| Ethyl acetate | VWR international | BJGC100-4 | |
| Hydroxlamine hydrochloride | Fisher Scientific | H330-100 | |
| Pyridine | Fisher Scientific | P368-500 | |
| Acetic anhydride | Fisher Scientific | A10-100 | |
| Dichloromethane (Methylene chloride) | Fisher Scientific | D37-500 | |
| Hexane | Fisher Scientific | H303-4 | |
| Hydrochloric acid (6M) | Fisher Scientific | S456-4 | |
| Hydroxylamine hydrochloride-15N | Icon services | IN5280 | |
| Acetic anhydride-2H (D6C4O3) | Acros Organics | AC174670050 | |
| D-glucose-U-13C | Cambridge Isotope Laboratories | CLM-1396-1 | |
| Ammonium sufate-15N | Cambridge Isotope Laboratories | NLM-713-1 | |
| Rapid-Vap | Labconco Corp. | 790002 | |
| Vacum pump | KNF Neuberger | D-79112 | |
| Hydrolysis flask | Fisher Scientific | 06 423A | |
| Derivatization microvial | Fisher Scientific | 06-100E | |
| GC | Hewlett-Packard | 6890 | |
| MS | Hewlett-Packard | 5972 | |
| LC-ESI-TOF-MS | Agilent Technologies | An Agilent 1200 series HPLC system coupled to an Agilent LC/MSD-TOF |
Odniesienia
- Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biology and Fertility of Soils. 29, 111-129 (1999).
- Kirk, J. L. Methods of studying soil microbial diversity. Journal of Microbiological Methods. 58, 169-188 (2004).
- Joergensen, R. G., Emmerling, C. Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils. Journal of Plant Nutrition and Soil Science. 169, 295-309 (2006).
- Guggenberger, G., Frey, S. D., Six, J., Paustian, K., Elliott, E. T. Bacterial and fungal cell-wall residues in conventional and no-tillage agroecosystems. Soil Science Society of America Journal. 63, 1188-1198 (1999).
- Amelung, W. Assessment Methods for Soil Carbon. Lal, R., Kimble, J. M., Follett, R. F., Stewart, B. A. , CRC/Lewis Publishers. Boca Raton, FL. 233-270 (2001).
- Guerrant, G. O., Moss, C. W. Determination of monosaccharides as aldononitrile, O-methyloxime, alditol, and cyclitol acetate derivatives by gas-chromatography. Analytical Chemistry. 56, 633-638 (1984).
- Zhang, X., Amelung, W. Gas chromatographic determination of muramic acid, glucosamine, mannosamine, and galactosamine in soils. Soil Biology and Biochemistry. 28, 1201-1206 (1996).
- Liang, C. Investigation of the molecular ion structure for aldononitrile acetate derivatized muramic acid. Journal of Microbiological Methods. 86, 224-230 (2011).
- He, H., Xie, H., Zhang, X. A novel GC/MS technique to assess 15N and 13C incorporation into soil amino sugars. Soil Biology and Biochemistry. 38, 1083-1091 (2006).
- Glaser, B., Gross, S. Compound-specific delta 13C analysis of individual amino sugars - a tool to quantify timing and amount of soil microbial residue stabilization. Rapid Communications in Mass Spectrometry. 19, 1409-1416 (2005).
- Liang, C., Balser, T. C. Mass spectrometric characterization of amino sugar aldononitrile acetate derivatives used for isotope enrichment assessment of microbial residues. Soil Biology and Biochemistry. 42, 904-909 (2010).
- Liang, C., Pedersen, J. A., Balser, T. C. Aminoglycoside antibiotics may interfere with microbial amino sugar analysis. Journal of Chromatography A. 1216, 5296-5301 (2009).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone