Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
A Semi-High-Throughput Adaptation of the NADH-Coupled ATPase Assay for Screening Small Molecule Inhibitors
W tym Artykule
Podsumowanie
A nicotinamide adenine dinucleotide (NADH)-coupled ATPase assay has been adapted to semihigh throughput screening of small molecule myosin inhibitors. This kinetic assay is run in a 384-well microplate format with total reaction volumes of only 20 µL per well. The platform should be applicable to virtually any ADP producing enzyme.
Streszczenie
ATPase enzymes utilize the free energy stored in adenosine triphosphate to catalyze a wide variety of endergonic biochemical processes in vivo that would not occur spontaneously. These proteins are crucial for essentially all aspects of cellular life, including metabolism, cell division, responses to environmental changes and movement. The protocol presented here describes a nicotinamide adenine dinucleotide (NADH)-coupled ATPase assay that has been adapted to semi-high throughput screening of small molecule ATPase inhibitors. The assay has been applied to cardiac and skeletal muscle myosin II's, two actin-based molecular motor ATPases, as a proof of principle. The hydrolysis of ATP is coupled to the oxidation of NADH by enzymatic reactions in the assay. First, the ADP generated by the ATPase is regenerated to ATP by pyruvate kinase (PK). PK catalyzes the transition of phosphoenolpyruvate (PEP) to pyruvate in parallel. Subsequently, pyruvate is reduced to lactate by lactate dehydrogenase (LDH), which catalyzes the oxidation of NADH in parallel. Thus, the decrease in ATP concentration is directly correlated to the decrease in NADH concentration, which is followed by change to the intrinsic fluorescence of NADH. As long as PEP is available in the reaction system, the ADP concentration remains very low, avoiding inhibition of the ATPase enzyme by its own product. Moreover, the ATP concentration remains nearly constant, yielding linear time courses. The fluorescence is monitored continuously, which allows for easy estimation of the quality of data and helps to filter out potential artifacts (e.g., arising from compound precipitation or thermal changes).
Wprowadzenie
Myosins are mechanochemical energy transducers that hydrolyze adenosine triphosphate (ATP) to generate directional movement along the filaments of the actin cytoskeleton in eukaryotes1,2. They have both structurally and kinetically adapted to their various intracellular functions, such as the transport of organelles, muscle contraction or the generation of cytoskeletal tension1,2. The myosin superfamily is represented by ~40 myosin genes belonging to ~12 distinct myosin classes in the human genome3,4. Members of the myosin classes play various roles in a highly diverse set of disorders, such as several cancers, neurological disorders, skeletal myopathies, and hypertrophic cardiomyopathy5,6. Given the large number of physiological and pathological functions of these molecular motors, it is not surprising that they are becoming increasingly recognized as drug targets for a variety of conditions7. Significant progress has been made recently in the discovery of new myosin inhibitors8,9,10 and activators11, and to improve the properties of existing ones12,13,14,15.
The nicotinamide adenine dinucleotide (NADH)-coupled ATPase assay has long been used to measure the ATPase activity of various enzymes, such as the sarcoplasmic reticulum Ca2+ pump ATPase16, the DNA repair ATPase Rad5417, the AAA+ ATPase p9718 or the microtubule motor kinesin19. The assay employs an ATP regeneration cycle. The adenosine diphosphate (ADP) generated by the ATPase is regenerated to ATP by pyruvate kinase (PK), which transforms one molecule of phosphoenolpyruvate (PEP) to pyruvate in parallel. Subsequently, pyruvate is reduced to lactate by lactate dehydrogenase (LDH). That, in turn, oxidizes one molecule of NADH to NAD. Therefore, the decrease in NADH concentration as a function of time equals the ATP hydrolysis rate. The ATP regeneration cycle keeps the ATP concentration nearly constant and the ADP concentration low as long as PEP is available. This results in linear time courses, making it simple to determine the initial reaction rates and helps to avoid product inhibition by ADP19. Although the NADH-coupled ATPase assay has already been adapted to a 96-well format20, the high reaction volumes (~150 µL) make it relatively expensive due to the high demand of reagents, rendering it less amenable to rapid screening of large numbers of compounds. Alternative methods, such as the malachite green assay19,21, which relies on the detection of the phosphate produced by the ATPase enzyme, were proven more suitable for miniaturization and high-throughput screening22,23,24. However, an endpoint assay is more likely to be affected by several artifacts (discussed below), which may remain undiscovered in the absence of full-time courses.
Here, the NADH-coupled ATPase assay has been optimized for semi-high throughput screening of small molecule inhibitors. Skeletal and cardiac muscle myosin II's and the myosin inhibitors blebbistatin8, para-aminoblebbistatin13 and para-nitroblebbistatin12 are used to demonstrate the power of the assay, which relies on NADH fluorescence as a readout. This protocol is amenable to screening projects focused on any ADP producing enzymes.
Protokół
1. Preparing stock solutions and reagents
- Prepare dithiothreitol (DTT) stock solution by dissolving crystalline DTT in distilled water to a final concentration of 1000 mM. Adjust the pH to 7.0 with 1 M NaOH solution. Aliquot and store at -20 °C.
- Prepare ATP stock solution by dissolving crystalline ATP in distilled water to a final concentration of 100 mM. Adjust the pH to 7.0 with 1 M NaOH solution. Aliquot and store at -20 °C.
- Prepare 10x NADH buffer containing 70 mM 3-(N-morpholino)propanesulfonic acid (MOPS), 10 mM MgCl2, 0.9 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), and 3 mM NaN3. Adjust the pH to 7.0 with 1 M NaOH solution. Store at 4 °C.
- Prepare 1x myosin buffer containing 10 mM MOPS and 0.1 mM EGTA. Adjust the pH to 7.0 with 1 M NaOH solution. Store at 4 °C. Add bovine serum albumin (BSA) and DTT to a final concentration of 0.1% (w/v%) and 1 mM, respectively, before use.
- Prepare 1x actin buffer containing 4 mM MOPS, 0.1 mM EGTA, 2 mM MgCl2, and 3 mM NaN3. Adjust the pH to 7.0 with 1 M NaOH solution. Store at 4 °C. Add BSA and DTT to a final concentration of 0.1% (w/v%) and 1 mM, respectively, before use.
- Prepare NADH stock solution by dissolving crystalline NADH in 10x NADH buffer to a final concentration of 5.5 mM. Aliquot and store at -20 °C.
- Prepare PEP stock solution by dissolving crystalline PEP in 10x NADH buffer to a final concentration of 50 mM. Aliquot and store at -20 °C.
- Prepare LDH stock solution by dissolving lyophilized LDH powder in a mixture of glycerol and 10x NADH buffer (50%:50%) to a final concentration of 2000 U/mL. Centrifuge the solution to remove any undissolved protein present (7,197 x g, 20 °C, 10 min). Transfer the supernatant into a clean centrifuge tube carefully. Aliquot and store at -20 °C.
- Prepare PK stock solution by dissolving lyophilized PK powder in a mixture of glycerol and 10x NADH buffer (50%:50%) to a final concentration of 10000 U/mL. Centrifuge the solution to remove any undissolved protein present (7,197 x g, 20 °C, 10 min). Transfer the supernatant into a clean centrifuge tube carefully. Aliquot and store at -20 °C.
- Reconstitute the lyophilized cardiac and skeletal muscle myosin II samples by adding 100 µL distilled water to obtain 10 mg/mL stock solutions corresponding to ~37.9 µM and ~40.8 µM myosin concentrations (monomeric), respectively. For further details, see manufacturer's instructions.
- Prepare F-actin from rabbit muscle acetone powder as described by Pardee and Spudich25.
2. Measuring ATPase activities and inhibitory effects of small molecule inhibitors
- Prepare compound plate.
- Dissolve compounds of interest in high-quality dimethylsulfoxide (DMSO).
- Create fifteen-step serial 1:2 dilutions starting from 10 mM compound concentration in DMSO.
- Transfer the samples to a 384-well polypropylene plate in triplicates (12.5 µL each) using a multichannel pipette. Use two rows on the compound plate for one compound (instead of three columns) to minimize the number of wells potentially affected by edge effects. Use the last three wells in the second row for each compound as negative control (DMSO only). Do not use the first and the last row on the plate for compound dilutions.
- Transfer pure DMSO into the wells of the first row (reserved for NADH calibration).
- Use the last row for positive control.
NOTE: Para-aminoblebbistatin at 4 mM concentration in DMSO was used here.
- Prepare 4500 µL of 20 µM diluted actin solution for each assay plate (384-well black-wall polystyrene microplate) by diluting actin stock solution in actin buffer. Mix the solution thoroughly by pipetting up and down 30x using a 5 mL pipette to reduce viscosity and heterogeneity by breaking actin filaments. Centrifuge the solution to remove any precipitated protein present (7,197 x g, 20 °C, 10 min). Carefully transfer the supernatant into a clean centrifuge tube.
- Prepare master mix containing LDH and PK enzymes ("enzyme mix"). For each assay plate, combine 171.4 µL of LDH solution, 171.4 µL of PK solution and 3189.3 µL or 3252.9 µL of myosin buffer for assays involving cardiac or skeletal muscle myosin II's, respectively, in a 15 mL conical centrifuge tube. Do not add any myosin at this point to avoid aggregation and precipitation.
- Prepare master mix containing all substrates ("substrate mix"). For each plate, combine 162.1 µL of ATP, 162.1 µL of PEP and 324.1 µL of NADH solution in a 15 mL conical centrifuge tube. Do not add actin at this point to avoid aggregation and precipitation.
- Create seven-step serial 1:2 dilutions of NADH for calibration starting from 250 µM.
- Mix 12.3 µL of NADH stock solution with 257.7 µL of myosin buffer in a 1.5 mL microcentrifuge tube.
- Aliquot 135 µL of myosin buffer into seven 1.5 mL microcentrifuge tubes.
- Transfer 135 µL of solution from the first tube into the second and mix by pipetting. Repeat until reaching the 7th tube.
- Use the last tube as no-NADH control (buffer only).
- Using an 8-channel pipette, transfer 20 µL of the NADH calibration solutions into the first row of the assay plate in triplicates.
- Add 68 µL of cardiac or 4.2 µL of skeletal muscle myosin II to the enzyme mix. Vortex briefly.
- Except the first row, dispense 8.4 µL of the prepared myosin-enzyme mix into each well of the assay plate using an automated dispenser.
- Transfer 100 nL of solutions from the compound plate to the assay plate containing enzyme mix using an automated liquid handling system equipped with a 100 nL pin tool head.
- Shake the assay plate for 1 min at room temperature at 1200 rpm using a microplate shaker.
- Add 4,052 µL of the centrifuged actin solution to the substrate mix. Vortex briefly.
- Dispense 11.6 µL of actin-substrate mix into each well of the assay plate (except first row) to start the enzymatic reaction using an automated dispenser.
- Shake the assay plate for 1 min at room temperature at 1200 rpm using a microplate shaker.
- Centrifuge the assay plate at 101 x g for 30 s.
- Make sure that the inner temperature of the plate reader has been stabilized at 25 ˚C. Load the plate and shake for another 30 s. This shaking step is necessary to make the shape of the liquid surface similar in each well and allows time for the plate to reach measurement temperature.
- Record NADH fluorescence for 30 min scanning the plate in 45 s intervals. Use a 380 nm, 10 nm bandwidth excitation filter and a 470 nm, 24 nm bandwidth emission filter in conjunction with a 425 nm cut-off dichroic mirror. Run the measurement in high-concentration mode. Optimize the number of flashes, detector gain, plate dimensions and measurement height before running the assays.
NOTE: Final assay conditions are 300 nM cardiac/20 nM skeletal muscle myosin II, 10 µM actin, 40 U/mL LDH, 200 U/mL PK, 220 µM NADH, 1 mM PEP, 1 mM ATP in a buffer containing 10 mM MOPS (pH = 7.0), 2 mM MgCl2, 0.15 mM EGTA, 0.1 mg/mL BSA, 0.5% (v/v) DMSO and 1 mM DTT. The total volume is 20 µL/well. The highest final compound concentration is 50 µM. 20 µM para-aminoblebbistatin in 0.5% DMSO serves as the positive control and 0.5% DMSO alone is the negative control. All measurements are carried out in triplicates.
3. Analyzing data
- Plot the observed fluorescence intensity against time for each well.
- Perform simple linear regression to determine the slope and intercept of the fluorescence responses for each well. The slope is proportional to the ATP (NADH) consumption rate, while the intercept is proportional to the NADH concentration at the beginning of the measurement (t = 0 s).
- Construct a calibration curve for NADH by plotting the intercepts obtained for the first row of the plate against the concentration of NADH. Make sure that the intercepts depend linearly on the NADH concentration.
NOTE: The intercepts estimate the real fluorescence intensities at t = 0 s with much more confidence than the average of the raw fluorescence intensity reads at t ≈ 0 s. - Perform simple linear regression to obtain the slope and intercept of the NADH calibration line.
NOTE: The intercept describes the fluorescence background signal (no NADH present), while the slope corresponds to the extrapolated/theoretical fluorescence intensity of a 1 M NADH solution in that particular experiment. - Divide the slope of the fluorescence response obtained for the rest of the wells by the slope of the NADH calibration line to convert fluorescence changes to ATP consumption rates.
- Plot the ATP consumption rates against the concentration of the inhibitor.
- To determine inhibitory constants, use appropriate statistical software to fit the dose-response data to the following quadratic equation corresponding to a simple one-to-one binding equilibrium model:
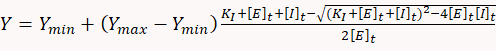
where Y is the ATP consumption rate, Ymin is the ATP consumption rate int the absence of inhibitor, Ymax is the theoretical ATP consumption rate at 100% inhibition, KI is the inhibitory constant, [E]t and [I]t are the total concentration of the enzyme (myosin) and inhibitor, respectively.
Wyniki
The typical plate layout map used for screening experiments is shown in Figure 1. The first and last rows are reserved for NADH calibration and positive control (20 µM para-aminoblebbistatin, 0.5% DMSO), respectively. The remaining rows (B to O) are used to test the inhibitory activity of compounds. Here, fifteen-step serial 1:2 dilutions starting from 10 mM compound concentration in DMSO are prepared and transferred from the compound plate to t...
Dyskusje
Critical steps in the protocol
Optimize plate layout by running several plates with negative control only (ATPase reaction with no inhibitor). Inspect the results carefully for patterns in reaction rates. For example, these may arise from edge effects and/or imperfections in the hydrophilic surface coating of "non-binding" plates. If a pattern is observed, change plate type and/or plate layout to minimize the artifacts. For example, a typical dose-response curve (16 co...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work was supported by a grant from the National Institute of Neurological Disorders and Stroke and National Institute on Drug Abuse NS096833 (CAM).
Materiały
| Name | Company | Catalog Number | Comments |
| 384-well Low Flange Black Flat Bottom Polystyrene NBS Microplate | Corning | 3575 | |
| ATP (Adenosine 5′-triphosphate disodium salt hydrate) | Sigma | A7699 | |
| Aurora FRD-IB Dispenser | Aurora Discovery, Inc. | 00017425 | |
| Biomek NXP Multichannel Laboratory Automation Workstation | Beckman Coulter | A31841 | |
| Blebbistatin | AMRI | N/A | Custom synthesis |
| BSA (Bovine Serum Albumin, Protease-Free) | Akron Biotech | AK1391 | |
| Centrifuge 5430 R, refrigerated, with Rotor FA-35-6-30 | Eppendorf | 022620663 | |
| Centrifuge 5430, non-refrigerated, with Rotor A-2-MTP | Eppendorf | 022620568 | |
| DMSO (Dimethyl sulfoxide) | Sigma | D2650 | |
| DTT (DL-Dithiothreitol) | Sigma | D5545 | |
| E1 ClipTip Multichannel Pipette; 384-format; 8-channel | Thermo Scientific | 4672010 | |
| E1 ClipTip Multichannel Pipette; 96-format; 8-channel | Thermo Scientific | 4672080 | |
| EGTA (Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid) | Sigma | E3889 | |
| EnVision 2104 Multilabel Plate Reader | PerkinElmer | 2104-0010 | |
| Glycerol | Sigma | G2025 | |
| LDH (L-Lactic Dehydrogenase from rabbit muscle) | Sigma | L1254 | |
| MgCl2.6H2O (Magnesium chloride hexahydrate) | Sigma | M2670 | |
| Microplate Shaker | VWR | 12620-926 | |
| Microplate, 384 well, PP, Small Volume, Deep Well, Natural | Greiner Bio-One | 784201 | |
| MOPS (3-(N-Morpholino)propanesulfonic acid) | Sigma | M1254 | |
| Myosin Motor Protein (full length) (Bovine cardiac muscle) | Cytoskeleton | MY03 | |
| Myosin Motor Protein (full length) (Rabbit skeletal muscle) | Cytoskeleton | MY02 | |
| NADH (β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate) | Sigma | N8129 | |
| NaN3 (Sodium azide) | Sigma | 71289 | |
| NaOH (Sodium hydroxide) | Sigma | S8045 | |
| Optical Filter CFP 470/24nm (Emission) | PerkinElmer | 2100-5850 | Barcode 240 |
| Optical Filter Fura2 380/10nm (Excitation) | PerkinElmer | 2100-5390 | Barcode 112 |
| Optical Module: Beta Lactamase | PerkinElmer | 2100-4270 | Barcode 418 |
| OriginPro 2017 software | OriginLab | N/A | |
| para-Aminoblebbistatin | AMRI | N/A | Custom synthesis |
| para-Nitroblebbistatin | AMRI | N/A | Custom synthesis |
| PEP (Phospho(enol)pyruvic acid monopotassium salt) | Sigma | P7127 | |
| PK (Pyruvate Kinase from rabbit muscle) | Sigma | P9136 | |
| Rabbit Muscle Acetone Powder | Pel Freez Biologicals | 41995-2 |
Odniesienia
- Heissler, S. M., Sellers, J. R. Kinetic Adaptations of Myosins for Their Diverse Cellular Functions. Traffic. 17 (8), 839-859 (2016).
- Hartman, M. A., Spudich, J. A. The myosin superfamily at a glance. Journal of Cell Science. 125 (Pt 7), 1627-1632 (2012).
- Berg, J. S., Powell, B. C., Cheney, R. E. A millennial myosin census. Molecular Biology of the Cell. 12 (4), 780-794 (2001).
- Sebe-Pedros, A., Grau-Bove, X., Richards, T. A., Ruiz-Trillo, I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biology and Evolution. 6 (2), 290-305 (2014).
- Newell-Litwa, K. A., Horwitz, R., Lamers, M. L. Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Disease Models & Mechanisms. 8 (12), 1495-1515 (2015).
- He, Y. M., Gu, M. M. Research progress of myosin heavy chain genes in human genetic diseases. Yi Chuan. 39 (10), 877-887 (2017).
- Rauscher, A. A., Gyimesi, M., Kovacs, M., Malnasi-Csizmadia, A. Targeting Myosin by Blebbistatin Derivatives: Optimization and Pharmacological Potential. Trends in Biochemical Sciences. 43 (9), 700-713 (2018).
- Straight, A. F., et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299 (5613), 1743-1747 (2003).
- Sirigu, S., et al. Highly selective inhibition of myosin motors provides the basis of potential therapeutic application. Proceedings of the National Academy of Sciences of the United States of America. 113 (47), E7448-E7455 (2016).
- Green, E. M., et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 351 (6273), 617-621 (2016).
- Morgan, B. P., et al. Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac Myosin. ACS Medicinal Chemistry Letters. 1 (9), 472-477 (2010).
- Kepiro, M., et al. para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor. Angewandte Chemie International Edition. 53 (31), 8211-8215 (2014).
- Varkuti, B. H., et al. A highly soluble, non-phototoxic, non-fluorescent blebbistatin derivative. Scientific Reports. 6, 26141 (2016).
- Verhasselt, S., et al. Discovery of (S)-3'-hydroxyblebbistatin and (S)-3'-aminoblebbistatin: polar myosin II inhibitors with superior research tool properties. Organic and Biomolecular Chemistry. 15 (9), 2104-2118 (2017).
- Verhasselt, S., Roman, B. I., Bracke, M. E., Stevens, C. V. Improved synthesis and comparative analysis of the tool properties of new and existing D-ring modified (S)-blebbistatin analogs. European Journal of Medicinal Chemistry. 136, 85-103 (2017).
- Warren, G. B., Toon, P. A., Birdsall, N. J., Lee, A. G., Metcalfe, J. C. Reconstitution of a calcium pump using defined membrane components. Proceedings of the National Academy of Sciences of the United States of America. 71 (3), 622-626 (1974).
- Kiianitsa, K., Solinger, J. A., Heyer, W. D. Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. Journal of Biological Chemistry. 277 (48), 46205-46215 (2002).
- Hanzelmann, P., Schindelin, H. Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97. Structure. 24 (1), 127-139 (2016).
- Hackney, D. D., Jiang, W. Assays for kinesin microtubule-stimulated ATPase activity. Methods in Molecular Biology. 164, 65-71 (2001).
- Kiianitsa, K., Solinger, J. A., Heyer, W. D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Analytical Biochemistry. 321 (2), 266-271 (2003).
- Carter, S. G., Karl, D. W. Inorganic phosphate assay with malachite green: an improvement and evaluation. Journal of Biochemical and Biophysical Methods. 7 (1), 7-13 (1982).
- Henkel, R. D., VandeBerg, J. L., Walsh, R. A. A microassay for ATPase. Analytical Biochemistry. 169 (2), 312-318 (1988).
- Rowlands, M. G., et al. High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Analytical Biochemistry. 327 (2), 176-183 (2004).
- Rule, C. S., Patrick, M., Sandkvist, M. Measuring In Vitro ATPase Activity for Enzymatic Characterization. Journal of Visualized Experiments. (114), 54305 (2016).
- Pardee, J. D., Spudich, J. A. Purification of muscle actin. Methods in Cell Biology. 24, 271-289 (1982).
- Zhang, J. H., Chung, T. D., Oldenburg, K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening. 4 (2), 67-73 (1999).
- Kovacs, M., Toth, J., Hetenyi, C., Malnasi-Csizmadia, A., Sellers, J. R. Mechanism of blebbistatin inhibition of myosin II. Chem Journal of Biological Chemistry. 279 (34), 35557-35563 (2004).
- Allingham, J. S., Smith, R., Rayment, I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nature Structural & Molecular Biology. 12 (4), 378-379 (2005).
- Kettlun, A. M., et al. Purification and Characterization of 2 Isoapyrases from Solanum-Tuberosum Var Ultimus. Phytochemistry. 31 (11), 3691-3696 (1992).
- Hulme, E. C., Trevethick, M. A. Ligand binding assays at equilibrium: validation and interpretation. British Journal of Pharmacology. 161 (6), 1219-1237 (2010).
- Motulsky, H. J., Neubig, R. R. Analyzing binding data. Current Protocols in Neuroscience. 52 (1), 7.5.1-7.5.65 (2010).
- Sehgal, P., Olesen, C., Moller, J. V. ATPase Activity Measurements by an Enzyme-Coupled Spectrophotometric Assay. Methods in Molecular Biology. 1377, 105-109 (2016).
- Solinger, J. A., Lutz, G., Sugiyama, T., Kowalczykowski, S. C., Heyer, W. D. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. Journal of Molecular Biology. 307 (5), 1207-1221 (2001).
- Banik, U., Roy, S. A continuous fluorimetric assay for ATPase activity. Biochemistry Journal. 266 (2), 611-614 (1990).
- Xiao, Y. X., Yang, W. X. KIFC1: a promising chemotherapy target for cancer treatment?. Oncotarget. 7 (30), 48656-48670 (2016).
- See, S. K., et al. Cytoplasmic Dynein Antagonists with Improved Potency and Isoform Selectivity. ACS Chemical Biology. 11 (1), 53-60 (2016).
- Datta, A., Brosh, R. M. New Insights Into DNA Helicases as Druggable Targets for Cancer Therapy. Frontiers in Molecular Biosciences. 5, 59 (2018).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone