Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Fabricating Multi-Component Lipid Nanotube Networks Using the Gliding Kinesin Motility Assay
W tym Artykule
Podsumowanie
This protocol describes a process for fabricating lipid nanotube networks using gliding kinesin motility in conjunction with giant unilamellar lipid vesicles.
Streszczenie
Lipid nanotube (LNT) networks represent an in vitro model system for studying molecular transport and lipid biophysics with relevance to the ubiquitous lipid tubules found in eukaryotic cells. However, in vivo LNTs are highly non-equilibrium structures that require chemical energy and molecular motors to be assembled, maintained, and reorganized. Furthermore, the composition of in vivo LNTs is complex, comprising of multiple different lipid species. Typical methods to extrude LNTs are both time- and labor-intensive, and they require optical tweezers, microbeads, and micropipettes to forcibly pull nanotubes from giant lipid vesicles. Presented here is a protocol for the gliding motility assay (GMA), in which large scale LNT networks are rapidly generated from giant unilamellar vesicles (GUVs) using kinesin-powered microtubule motility. Using this method, LNT networks are formed from a wide array of lipid formulations that mimic the complexity of biological LNTs, making them increasingly useful for in vitro studies of lipid biophysics and membrane-associated transport. Additionally, this method is capable of reliably producing LNT networks in a short time (<30 min) using commonly used laboratory equipment. LNT network characteristics such as length, width, and lipid partitioning are also tunable by altering the lipid composition of the GUVs used for fabricating the networks.
Wprowadzenie
The fabrication of lipid nanotube (LNT) networks is of increasing interest for in vitro examination of nonequilibrium lipid structures1,2,3. Cells use lipid tubules for the diffusive transport of proteins4 and nucleic acids5 as well as cell-to-cell communication6,7. The endoplasmic reticulum and Golgi apparatus are particularly interesting, as these membrane-bound organelles are the primary locations for lipid and protein synthesis as well as transport of these integral biomolecules within the cytoplasm of a cell8,9. The membranes of these organelles are comprised of multiple lipid species including sphingolipids, cholesterol, and phospholipids10 that ultimately help define their functionality. Thus, to more closely replicate and study these organelles, in vitro LNTs must be fabricated from vesicles with increasingly complex lipid formulations11.
Giant unilamellar vesicles (GUVs) are used pervasively for studying lipid membrane behavior because they can be reliably synthesized with complex formulations that include cholesterol, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI)12,13. Described here is a method to fabricate LNTs from GUVs with varying lipid formulations using the gliding motility assay (GMA), in which LNTs are extruded based on the work performed by kinesin motors and microtubule filaments acting on GUVs. In this system, kinesin motor proteins adsorbed to a surface propel biotinylated microtubules, converting chemical energy from the hydrolysis of ATP into useful work (specifically, the extrusion of LNTs from biotinylated vesicles)11. The resulting LNT network provides a model platform to study effects of the differences in lipid phases on changes in LNT morphology.
Briefly, kinesin motor proteins are introduced into a flow chamber in a solution containing casein, which enables the adsorption of the motors onto the glass surface of the chamber. Next, biotinylated microtubules in a solution containing ATP flow through the chamber and are allowed to bind to the kinesin motors and begin motility. A streptavidin solution is then introduced into the chamber and allowed to bind non-covalently to the microtubules. Finally, GUVs containing a biotinylated lipid are introduced into the chamber and bind to the streptavidin-coated microtubules, then extrude LNTs to form large-scale networks over the course of 15–30 min. This method produces large, branched LNT networks using standard laboratory equipment and reagents at a low cost11.
Access restricted. Please log in or start a trial to view this content.
Protokół
1. Preparation of stock microtubule solutions
CAUTION: Safety goggles, gloves and a lab coat should always be worn throughout the protocol.
- Prepare 5x BRB80 buffer: add 24.19 g of PIPES (piperazine-N,N′-bis[2-ethanesulfonic acid]) and 0.38 g of EGTA (ethylene glycol-bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid) to a 1 L glass bottle. Add 1 mL of 1 M MgCl2 and adjust the pH to 6.9 with KOH. Add deionized water to bring the solution to a 500 mL final volume.
- Prepare 100 mM stock of GTP solution: weigh 52 mg of GTP and suspend in 1 mL of distilled water. Divide 100 mM solution into 20 µL aliquots and store at -20 °C.
- Prepare GPEM solution: mix 200 µL of 5x BRB80, 10 µL of 100 mM GTP solution, 100 µL of 100% glycerol, and 600 µL of deionized water. Divide GPEM solution in 100 µL aliquots and store in -20 °C.
- Prepare microtubule solution by reconstituting vials of commercially available, lyophilized tubulin (one vial each of biotinylated, fluorescently labeled, and unlabeled) in cold (4 °C) GPEM solution to a stock concentration of 5 mg/mL.
- Perform microtubule polymerization by mixing 4 µL of biotinylated tubulin, 4 µL of fluorescently labeled tubulin, and 24 µL of unlabeled tubulin (all at 5 mg/mL concentrations) to create a ratio of 1:1:6 at a final volume of 32 mL. Keep it on ice. Divide the tubulin mixture into 2 µL aliquots and store at -80 °C until needed.
NOTE: Efficient polymerization requires that the concentration of tubulin is equal to or above the critical concentration (5 mg/mL)14. Here, selection of the tubulin ratio is optimized for a sufficient concentration biotinylated tubulin to efficiently bind streptavidin and GUVs, as well as a sufficient concentration of fluorescent tubulin for microscopic characterization.
2. Preparation of giant unilamellar vesicles (GUVs)
- Agarose film preparation
NOTE: This protocol is adapted from Greene et al.15.- Prepare a 1% w/v solution by mixing 1 g of agarose in 100 mL of deionized water in 250 mL Erlenmeyer flask. Use a standard microwave to heat the agarose solution for 1–2 min.
NOTE: The solution will become translucent once the agarose is completely dissolved. Allow the solution to cool to 65–75 °C before use. - Use a cut 1,000 µL pipette tip to pipette 300–400 µL of agarose solution onto a 25 mm x 25 mm glass coverslip. While holding the edge of the coverslip with gloved fingers, use another 1,000 µL pipette tip to spread the melted agarose evenly across the coverslip.
NOTE: Maintaining the agarose at 65–75 °C will allow for efficient spreading on the coverslip surface. - Dry the agarose-coated coverslips in a 37 °C incubator for at least 2 h, at which point the agarose will become transparent. Store the coverslips by placing the agarose coated surface facing upwards on a clean surface such as lint-free paper or wax-based film at room temperature (RT).
- Prepare a 1% w/v solution by mixing 1 g of agarose in 100 mL of deionized water in 250 mL Erlenmeyer flask. Use a standard microwave to heat the agarose solution for 1–2 min.
- Lipid formulation
- Retrieve lipids dissolved in chloroform from a -20 °C freezer and place them in a chemical fume hood until they reach RT.
- Calculate the volume of lipid stock of each component lipid needed for the formulation using following formula:
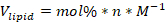
Where: Vlipid is the volume of lipid to use, mol % is the molar percentage of the lipid component, n is the total number of moles of lipid used in the formulation, and M is the concentration of lipid in molar units.
NOTE: For example, if using a formulation containing 45 mol% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) with a stock solution concentration of 12.72 mM, and 1 micromole of total lipids in the formulation, the volume of DOPC stock used in the formulation would be:
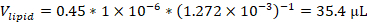
- Mix the lipids together at the calculated ratio in a glass vial in the chemical fume hood.
- Pipette 30 µL of lipid solution onto the agarose-coated coverslips on a preheated hot plate that is set above the melting point of the saturated lipid component of the formulation (e.g., 50 °C hot plate for a lipid with Tm of 40 °C).
- Spread the solution across the agarose film in a circular motion using the long edge of an 18 G needle to until the chloroform has evaporated and a uniform layer of lipid has formed. Hold the edge of the coverslip with gloved fingers while performing this step.
NOTE: Care should be taken not to damage the agarose layer with the needle. - Place the coverslip with agarose layer and lipid film in an aluminum foil-covered Petri dish, film side facing upwards, and place the Petri dish in a vacuum desiccator for at least 2 h to remove residual solvent.
- In the meantime, prepare 560 mM sucrose solution by mixing 1.92 g of sucrose with 10 mL of deionized water.
NOTE: The concentration of the sucrose solution depends on the osmolarity of the buffer GUVs are diluted in. Typically, the osmolarity of the sucrose solution should be no more than 10% larger than the buffer the GUVs will be diluted in, specifically the motility buffer (see step 3.11). - GUV formation
- Adhere an adhesive chamber to the coverslip coated with the lipid film by gently pressing the adhesive chamber onto the lipid-coated coverslip with the lipid film facing upwards, ensuring that a tight seal is formed.
- Add 400 µL of 560 mM sucrose solution (prepared in step 2.3) to the chamber.
- Place the coverslip into the humidity chamber and close the lid.
- Place the humidity chamber on a preheated hot plate set above the melting point of the saturated lipid component of the formulation.
- Allow vesicles to form for ≥1 h before recovery.
NOTE: Vesicle formation may be checked with fluorescence microscopy with a 40x air objective lens.
3. Preparation of motility assay stocks and reagents
- Preparation of casein stock
- Add 3 g of dry casein to a 50 mL conical centrifuge tube, then add 30 mL of 1x BRB80 and rotate until the solution becomes viscous. Centrifuge the tube at 15,000 x g for 30 min.
- Transfer the supernatant into another 50 mL conical centrifuge tube and discard the pellet. Filter the solution through a 1 µm syringe filter, collecting the solution in a 50 mL conical vial. Filter the solution through a 0.2 µm filter, collecting the solution in a 50 mL conical vial.
- Determine the protein concentration by measuring the absorbance at 280 nm using a UV-Vis spectrophotometer and quartz cuvette.
- Calculate the casein concentration in mg/mL using the following formula:
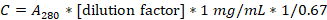
- Dilute the solution to 20 mg/mL in 1x BRB80, divide into 100 µL aliquots, and store at -20 °C.
- Prepare glucose oxidase (2 mg/mL) stock solution by mixing 2 mg of glucose oxidase with 1 mL of 1x BRB80. Divide into 100 µL aliquots and store at -20 °C.
- Prepare catalase (0.8 mg/mL) stock solution by mixing 0.8 mg of catalase with 1 mL of 1x BRB80. Divide into 100 µL aliquots and store at -20 °C.
- Prepare 2 M glucose solution by suspending 0.3 g of D-glucose in 1 mL of deionized water. Divide into 100 µL aliquots and store at -20 °C.
- Prepare 100 mM DTT stock by suspending 0.015 g of DTT in 1 mL of deionized water. Divide into 100 µL aliquots and store at -20 °C.
- Prepare 100 mM Mg-ATP stock by suspending 0.055 g of disodium ATP in 1 mL solution of 100 mM MgCl2. Divide into 100 µL aliquots and store at -20 °C.
- Prepare 100 mM Mg-AMP-PNP solution by suspending out 0.055 g of AMP-PNP in a 1 mL solution of 100 mM MgCl2, then divide into 100 µL aliquots and store at -20 °C.
- Prepare 100 mM Trolox solution by adding 25.03 mg of Trolox to 1 mL of methanol and store at -20 °C.
- Prepare 10 mg/mL streptavidin solution by adding 1 mg of streptavidin to 100 µL of BRB80, then divide into 2 µL aliquots and and store at -80ۛ °C.
- Prepare BRB90CAT by mixing 200 µL of 5x BRB80, 20 µL of casein solution, 10 µL of MgATP solution, 10 µL of Trolox, 5 µL of paclitaxel solution, and 765 µL of DI water. Store at 4 °C.
- Prepare the motility solution by mixing 192 µL of BRB80CAT, 2 µL of D-glucose solution, 2 µL of glucose oxidase solution, 2 µL of DTT solution, and 2 µL of catalase solution. Store at 4 °C.
- Prepare a 1 µM kinesin solution by diluting the stock kinesin solution in BRB80CAT (e.g., 2 µL of 50 mM kinesin solution in 98 µL of BRB80CAT) and store at 4 °C.
- Prepare a 10 µg/mL microtubule solution by diluting 10 µL of stabilized microtubules in 90 µL of room temperature BRB80CAT. Store at RT.
- Prepare a 10 µg/mL streptavidin solution by adding 0.1 µL of 10 mg/mL streptavidin solution in 99.9 µL of motility solution. Store at 4 °C.
- Prepare a 12x GUV solution by diluting 5 µL of GUV stock into 55 µL of motility solution. Store at 4 °C.
- Polymerization of tubulin into microtubules
- Collect a previously prepared 2 µL aliquot of tubulin from the -80 °C freezer (prepared in step 1.5) and place into a 37 °C water bath for 30 min.
- Prepare a 2 mM paclitaxel solution by adding 1.71 mg of paclitaxel in 1 mL of anhydrous DMSO, divide into 10 µL aliquots, and store at -20 °C.
- Freshly prepare BRB80T by mixing 99.5 µL of 1x BRB80 with 0.5 µL of 2 mM paclitaxel. Warm 100 µL of BRB80T to 37 °C in the water bath.
- After 30 min, add the 100 µL of BRB80T to the tubulin aliquot to stabilize the microtubules. Store at RT.
4. Gliding motility assay (GMA)
- Prepare a flow chamber by affixing two strips of double-sided tape separated by 5 mm onto a glass slide. Repeat this process until three layers of tape comprise each strip.
- Place a coverslip on top of the tape, then press down gently on the coverslip/tape interface with a tweezer or pen to ensure sufficient adhesion.
NOTE: The channel should be 5 mm in width by 25 mm in length by 300 mm in height. - Pipette 30 µL of 1 mm kinesin solution (prepared in step 3.12) into the flow cell and let it incubate for 5 min.
NOTE: The casein forms a bilayer on the surface of the coverslip/glass slide and facilitates attachment of the kinesin tail to the surface. - Pipette 30 µL of 10 µg/mL microtubule solution (prepared in step 3.13) into the flow cell, using a laboratory wipe pressed gently against the opposite end of the flow channel to facilitate solution exchange. Incubate for 5 min.
- Wash the flow cell 1x–3x with 1x motility solution at RT (prepared in step 3.11).
NOTE: Fluorescence microscopy using a 40x air objective may be used at this point to confirm microtubule attachment and motility. Microtubules appear as fluorescent filaments (tens of microns in length) moving (gliding) across the surface at ~0.5 μm/s (Figure 1). - Pipette 30 µL of 10 µg/mL streptavidin solution (prepared in step 3.14) into the flow cell using a laboratory wipe pressed gently against the opposite end of the flow channel to facilitate solution exchange. Incubate for 10 min.
- Flow 30 µL of 12x GUV solution (prepared in step 3.15) into the flow using a laboratory wipe pressed gently against the opposite end of the flow channel to facilitate solution exchange. Incubate for 30 min.
- Add 2 µL of 100 mM AMP-PNP solution (prepared in step 3.7) to stop motility, then seal the chamber with sealant.
5. LNT network characterization
- Transfer the flow chamber to an inverted microscope for imaging.
- Choose the appropriate filter set based on the wavelengths of the fluorescent lipids or tubulin used. For example, when using Texas Red-labeled lipids, use a 560 nm/25 nm excitation filter and 607 nm/36 nm emission filter.
- Use a 100x oil objective to focus on the surface of the coverslip.
- Image the LNT networks using fluorescence microscopy.
NOTE: LNTs are linear structures extruding from the larger vesicles. LNTs much smaller than GUVs and have weaker fluorescence signals. Thus, the exposure and contrast must be adjusted accordingly to image LNTs. These adjustments also lead to overexposure of the GUVs, and thus, it is recommended that the lamellarity and phase separation in GUVs be characterized independently. - Focus the microscope on a network of interest and take a standard or tiled image.
- Adjust the exposure time and neutral density filters to image the LNTs and minimize saturated exposure of the GUVs. Acquire images both in red and green channels.
NOTE: Here, the red channel permits visualization of the Texas-Red lipids, while the green channel permits visualization of the microtubules (e.g., Oregon Green lipids and HiLyte488 dyes). - Create a composite image by overlaying the red and green channels (Figure 1).
- Characterization of LNT networks by measuring LNT length
- Open the acquired images using an image analysis software such as ImageJ.
- Calibrate the scale for the microscope by using the set scale feature, fill in the pixels to the mm conversion factor, and click OK.
NOTE: The conversion factor is dependent on the microscope, objective lens, and camera, and it can be obtained using a microscope calibration slide. It is generally expressed in pixels/mm. - Use the Multipoint line tool to measure the length of the nanotubes starting from the parent GUV. Hold Ctrl +M to measure the length.
- Continue measuring the lengths of individual tubes following the steps above. The image processing tool will save each new measurement in the results window.
- Hold Ctrl + D after drawing each line to keep track of which tubes have been measured.
- Measuring LNT thickness (Figure 2)
- Open the image in ImageJ and select the Threshold feature under the Image tab.
- Click Apply to apply the threshold.
- Draw a rectangle of known length over the desired tube (black pixels have a value of 0, and red pixels have a value of 255).
- Measure the integrated density of the area.
- Divide the density by the length (in pixels) of the LNT to obtain the thickness (in pixels).
NOTE: The thickness measurements may only be compared across images when the imaging settings and threshold are set identically.
- Determining the lipid partitioning in nodes of LNTs (Figure 3)
- Open the image in ImageJ.
- Use the Line tool to draw a line over the desired node.
- Measure the node intensity in both the Oregon Green and Texas Red channels.
- Move the line to the LNT and measure the LNT intensity in both the Oregon Green and Texas Red channels.
Access restricted. Please log in or start a trial to view this content.
Wyniki
LNT networks (Figure 4) were fabricated using the described protocol, which uses the work performed by kinesin transport of microtubules to extrude LNTs from GUVs. Briefly, GUVs were prepared using agarose gel rehydration using sucrose solution, and microtubules were polymerized in GPEM solution and stabilized in BRB80T. Next, kinesin motors were introduced into a flow cell forming an active layer of motors on the surface of the coverslip. Microtubules were then introduced and a strepta...
Access restricted. Please log in or start a trial to view this content.
Dyskusje
LNT networks are a useful tool for in vitro studies to membrane properties and the transport of biomolecules such as transmembrane proteins. Moreover, using complex lipid formulations to fabricate LNT networks enables more biologically relevant studies. Other fabrication studies have used either 1) simple lipid formulations and multilamellar vesicles or 2) more cumbersome motility techniques to fabricate networks from GUVs comprised of complex lipid formulations. The method described here enables the efficient fabricatio...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
Sandia National Laboratories is a multi-mission laboratory managed and operated by National Technology & Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. DOE’s National Nuclear Security Administration under contract DE-NA-0003525. This paper describes objective technical results and analysis. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the U.S. Department of Energy or the United States Government.
Podziękowania
This work was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering (BES-MSE). Kinesin synthesis and fluorescence microscopy were performed through a user project (ZIM) at the Center for Integrated Nanotechnologies, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| 100x/1.4 Numerical Aperture Oil Immersion Objective | Olympus | 1-U2B836 | Olympus UPlanSApo 100x/1.40 Oil Objective Infinity Corrected, RMS Thread Working Distance 0.12mm |
| 3.0 ND Filter | Olympus | Neutral Density Filter | |
| AMP-PNP | Sigma-Aldrich | A2647 | (β,γ-imidoadenosine 5′-triphosphate) |
| ATP | Sigma-Aldrich | A7699 | Adenosine 5'-triphosphate disodium salt hydrate BioXtra |
| Brightline Pinkel DA/FI/TR/Cy5/Cy7-5X-A000 filter set | Semrock | LED-DA/FI/TR/Cy5/Cy7-5X-A-000 | BrightLine Pinkel filter set, optimized for DAPI, FITC, TRITC, Cy5 & Cy7 and other like fluorophores, illuminated with LED-based light sources |
| Casein | Sigma-Aldrich | 22090 | Casein hydrolysate for microbiology |
| Catalase | Sigma-Aldrich | C9322 | Catalase from Bovine Liver |
| Chloroform | Sigma-Aldrich | 288306 | Chloroform anhydrous contains 0.5-1.0% ethanol as stabilizer |
| Cholesterol | Avanti | 700000P | cholesterol (ovine wool, >98%) (powder) |
| D-Glucose | Sigma-Aldrich | G7021 | D-(+)-Glucose powder, BioReagent, suitable for cell culture, suitable for insect cell culture, suitable for plant cell culture, ≥99.5% |
| DOPC | Avanti | 850375C | 1,2-Dioleoyl-sn-glycero-3-phosphocholine (in chloroform) |
| DOPE-Biotin | Avanti | 870282C | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (sodium salt) |
| DPPC | Avanti | 850355P | 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (powder) |
| DPPE-Biotin | Avanti | 870285P | 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (sodium salt) |
| DTT | Sigma-Aldrich | 43816 | DL-Dithiothreitol solution 1 M |
| EGTA | Sigma-Aldrich | E4378 | EGTA, Egtazic acid, Ethylene-bis(oxyethylenenitrilo)tetraacetic acid, Glycol ether diamine tetraacetic acid |
| Glucose Oxidase | Sigma-Aldrich | G6125 | Glucose Oxidase from Aspergillus niger Type II, ≥10,000 units/g solid (without added oxygen) |
| Glycerol | Fisher | G33 | Glycerol (Certified ACS), Fisher Chemical |
| GTP | Sigma-Aldrich | G8877 | Guanosine 5′-triphosphate sodium salt hydrate |
| IX-81 Olympus Microscope | Olympus | N/A | IX81 Inverted Microscope from Olympus |
| KOH | Sigma-Aldrich | 1050121000 | Potassium Hydroxide |
| Magnesium Chloride | Sigma-Aldrich | M1028 | 1.00 M magnesium chloride solution |
| Orca Flash 4.0 Digital Camera | Hamamatsu | C13440-20CU | ORCA-Flash 4.0 V3 Digital CMOS camera |
| Oregon Green-DHPE | Invitrogen | O12650 | Oregon Green 488 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine |
| Paclitaxel | ThermoFisher | P3456 | Paclitaxel (Taxol Equivalent) - for use in research only |
| PIPES | Sigma-Aldrich | P6757 | 1,4-Piperazinediethanesulfonic acid, Piperazine-1,4-bis(2-ethanesulfonic acid), Piperazine-N,N′-bis(2-ethanesulfonic acid) |
| Texas Red-DHPE | Invitrogen | T1395MP | Texas Red 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine, Triethylammonium Salt |
| Trolox | Sigma-Aldrich | 238813 | (±)-6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid |
| Tubulin, Biotin | Cytoskeleton | T333P | Tubulin protein (biotin) porcine brain |
| Tubulin, Hy-Lite 488 | Cytoskeleton | TL488M | Tubulin protein (fluorescent HiLyte 488) porcine brain |
| Tubulin, Unlabeled | Cytoskeleton | T240 | Tubulin protein porcine brain |
Odniesienia
- Bouxsein, N. F., Carroll-Portillo, A., Bachand, M., Sasaki, D. Y., Bachand, G. D. A continuous network of lipid nanotubes fabricated from the gliding motility of kinesin powered microtubule filaments. Langmuir. 29 (9), 2992-2999 (2013).
- Paxton, W. F., Bouxsein, N. F., Henderson, I. M., Gomez, A., Bachand, G. D. Dynamic assembly of polymer nanotube networks via kinesin powered microtubule filaments. Nanoscale. 7 (25), 10998-11004 (2015).
- Leduc, C., et al. Cooperative extraction of membrane nanotubes by molecular motors. Proceedings of the National Academy of Sciences of the United States of America. 101 (49), 17096-17101 (2004).
- Lippincott-Schwartz, J., Roberts, T. H., Hirschberg, K. Secretory protein trafficking and organelle dynamics in living cells. Annual Review of Cell and Developmental Biology. 16, 557-589 (2000).
- Belting, M., Wittrup, A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. Journal of Cell Biology. 183 (7), 1187-1191 (2008).
- Rustom, A., Saffrich, R., Markovic, I., Walther, P., Gerdes, H. H. Nanotubular highways for intercellular organelle transport. Science. 303 (5660), 1007-1010 (2004).
- Onfelt, B., Nedvetzki, S., Yanagi, K., Davis, D. M. Cutting edge: Membrane nanotubes connect immune cells. Journal of Immunology. 173 (3), 1511-1513 (2004).
- Sciaky, N., et al. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 139 (5), 1137-1155 (1997).
- Sprong, H., van der Sluijs, P., van Meer, G. How proteins move lipids and lipids move proteins. Nature Reviews Molecular Cell Biology. 2 (7), 504-513 (2001).
- Keenan, T. W., Morre, D. J. Phospholipid class and fatty acid composition of golgi apparatus isolated from rat liver and comparison with other cell fractions. Biochemistry. 9 (1), 19-25 (1970).
- Imam, Z. I., Bachand, G. D. Multicomponent and Multiphase Lipid Nanotubes Formed by Gliding Microtubule-Kinesin Motility and Phase-Separated Giant Unilamellar Vesicles. Langmuir. 35 (49), 16281-16289 (2019).
- Wesolowska, O., Michalak, K., Maniewska, J., Hendrich, A. B. Giant unilamellar vesicles - a perfect tool to visualize phase separation and lipid rafts in model systems. Acta Biochimica Polonica. 56 (1), 33-39 (2009).
- Momin, N., et al. Designing lipids for selective partitioning into liquid ordered membrane domains. Soft Matter. 11 (16), 3241-3250 (2015).
- Fygenson, D. K., Braun, E., Libchaber, A. Phase diagram of microtubules. Physical Review E. 50, 1579(1994).
- Greene, A. C., Sasaki, D. Y., Bachand, G. D. Forming Giant-sized Polymersomes Using Gel-assisted Rehydration. Journal of Visualized Experiments. (111), (2016).
- Bachand, M., et al. Directed self-assembly of 1D microtubule nano-arrays. Royal Society of Chemistry Advances. 4 (97), 54641-54649 (2014).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone