Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Effect of Hyaluronic Acid 35 kDa on an In Vitro Model of Preterm Small Intestinal Injury and Healing Using Enteroid-Derived Monolayers
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
This protocol describes a method to establish and perform a scratch wound assay on two-dimensional (2D) monolayers derived from three-dimensional (3D) enteroids isolated from non-human primate ileum.
Streszczenie
In vitro scratch wound assays are commonly used to investigate the mechanisms and characteristics of epithelial healing in a variety of tissue types. Here, we describe a protocol to generate a two-dimensional (2D) monolayer from three-dimensional (3D) non-human primate enteroids derived from intestinal crypts of the terminal ileum. These enteroid-derived monolayers were then utilized in an in vitro scratch wound assay to test the ability of hyaluronan 35 kDa (HA35), a human milk HA mimic, to promote cell migration and proliferation along the epithelial wound edge. After the monolayers were grown to confluency, they were manually scratched and treated with HA35 (50 µg/mL, 100 µg/mL, 200 µg/mL) or control (PBS). Cell migration and proliferation into the gap were imaged using a transmitted-light microscope equipped for live-cell imaging. Wound closure was quantified as percent wound healing using the Wound Healing Size Plugin in ImageJ. The scratch area and rate of cell migration and the percentage of wound closure were measured over 24 h. HA35 in vitro accelerates wound healing in small intestinal enteroid monolayers, likely through a combination of cell proliferation at the wound edge and migration to the wound area. These methods can potentially be used as a model to explore intestinal regeneration in the preterm human small intestine.
Wprowadzenie
Necrotizing enterocolitis (NEC) is one of the most common gastrointestinal emergencies in preterm infants1. The disease is characterized by severe intestinal inflammation that can rapidly deteriorate to intestinal necrosis, sepsis, and potentially death. Although the etiology is unclear, evidence suggests NEC is multifactorial and the result of a complex interaction of feeding, abnormal bacterial colonization, and an immature intestinal epithelium2,3. Preterm infants have increased intestinal permeability, abnormal bacterial colonization, and low enterocyte regenerative capacity4,5, increasing their risk for intestinal barrier dysfunction, bacterial translocation, and NEC development. Therefore, identifying strategies or interventions to accelerate intestinal epithelial maturation and promote regeneration or healing of the intestinal epithelium is critical in preventing this deadly disease.
Studies have demonstrated that human milk (HM) is protective against NEC in preterm infants6,7,8,9,10,11. Both human and animal studies have shown that bovine-based formula increases intestinal permeability and is directly toxic to intestinal epithelial cells2,12. Although not fully elucidated, evidence suggests the protective effects of HM are mediated through bioactive components such as lactoferrin, immunoglobulin A (IgA), and HM oligosaccharides13. HM is also rich in hyaluronan (HA), a uniquely nonsulfated glycosaminoglycan with repeating D-glucuronic acid and N-acetyl-D-glucosamine disaccharides14,15. Importantly, we have shown that oral 35 kDa HA (HA35), an HM HA mimic, attenuates the severity of intestinal injury, prevents bacterial translocation, and decreases mortality in a murine NEC-like intestinal injury model16,17.
Here, the effects of HA35 on intestinal healing and regeneration in vitro are further investigated. Currently, the most widely used in vitro assay for intestinal wounding and repair is a scratch wound assay performed in colorectal cancer (CRC) cell monolayers. The physiological relevance of such a model to the preterm infant intestine is limited, as wound repair of CRC cells relies heavily upon the highly proliferative nature of cancer cells rather than stem cell-driven repair processes18. To overcome this limitation, the establishment of a 2D enteroid scratch wound model, including the procedure of isolating and maintaining primary stem cell-derived small intestinal enteroids from preterm non-human primates (NHP), is described here. Given preterm NEC is most often reported in the distal small intestine, the use of primary epithelial cell organoids in a model of intestinal damage and repair provides a more physiologically translatable in vitro model compared with existing models utilizing traditional colorectal monolayers18,19.
Protokół
All animal procedures in this study were approved by the University of Oklahoma Health Sciences Center Institutional Animal Care and Use Committee. Following institutional approval, fetal small intestine convenience samples from a preterm non-human primate (NHP, 90% gestation, olive baboon, Papio anubis) were obtained following euthanasia for a separate study (Protocol #101523-16-039-I)20.
1. Establishment of preterm non-human primate 3D intestinal enteroids
- Media preparation
NOTE: Media can be prepared up to 1 week in advance of crypt isolation following standard aseptic technique. Penicillin/streptomycin (final concentration 1%) may be used in place of the broad-spectrum antibiotics for primary cells if desired.- Prepare human organoid growth media + Y-27632 (HOGMY) by mixing 50 mL of organoid growth medium human basal medium with 50 mL of organoid supplement (Table of Materials). Add 200 µL of the broad-spectrum antibiotics (final concentration 100 µg/mL) (Table of Materials) and Y-27632 (to a final concentration of 10 µM). Warm on the benchtop to room temperature.
- Prepare human organoid growth media (HOGM) by mixing 50 mL of organoid growth medium human basal medium with 50 mL of organoid supplement. Add 200 µL of the broad-spectrum antibiotics (100 µg/mL). Warm on the benchtop to room temperature.
NOTE: HOGM will be used when changing media beyond the first 2-3 days post-passage. - Chill 100 mL of 1x phosphate-buffered saline (PBS) without calcium or magnesium until ice-cold.
- Prepare 50 mL of wash buffer by combining 1x PBS without calcium or magnesium with 0.1% bovine serum albumin (BSA). Chill it until ice-cold.
- Prepare DMEM-F12 wash buffer by adding 1 mL of the broad-spectrum antibiotics (100 µg/mL) to 500 mL of DMEM/F12 (Dulbecco's Modified Eagle's Medium/Nutrient Ham's Mixture F12 + 15 mM HEPES buffer). Keep it ice-cold.
- Crypt isolation and plating
NOTE: Extracellular matrix (ECM)-based hydrogel must remain on the ice during the entire procedure. This procedure assumes enough tissue to populate six ECM-based hydrogel domes. If a different density of crypts is harvested, the dome number should be altered accordingly.- Thaw 150 µL of growth factor reduced (GFR) basement membrane extract (BME) (phenol red-free) overnight on the ice at 2-8 °C.
- Incubate a 24-well, flat-bottom, tissue culture-treated polystyrene plate in an incubator at 37 °C and 5% CO2 for a minimum of 30 min before use.
- Following NHP euthanasia and tissue collection, flush the ileum segment of debris with a 1,000 µL pipette tip using ice-cold 1x PBS in a disposable Petri dish.
NOTE: Fresh tissue must be harvested quickly and kept ice-cold to harvest healthy enteroids. - Cut the ileum longitudinally along the entire intestine, and wash with ice-cold PBS 3x. Mince the tissue into small fragments (approximately 2 mm in length) with sterile scissors over a 50 mL conical tube containing 15 mL ice-cold PBS, starting from the portion of the ileal segment closest to the tube.
- Use a 10 mL serological pipette to disrupt the crypts by pipetting up and down 5-10x. Let the tissue segments settle to the bottom of the tube, then remove the supernatant and replace it with fresh, ice-cold PBS. Repeat this process a minimum of 7x-10x until the supernatant is clear and free of debris.
- Once clear, remove the supernatant and resuspend the crypts in 25 mL of cell dissociation reagent. Place the tube on a rocking platform at 20 rpm for 15 min at room temperature.
- Remove the tube from the rocker, allow the cells to settle for approximately 30 s to 1 min, and remove the supernatant. Add 10 mL of ice-cold wash buffer. Pipette up and down with a p1000 pipette to further dissociate into single crypts.
- Filter the tissue through a 70 µm cell strainer into a new 50 mL conical tube. Rinse the original 50 mL conical tube with 10 mL of ice-cold wash buffer and filter through the 70 µm strainer to ensure all the crypts have been removed from the original tube. Repeat this washing of tissue for a total of 4x rinses.
- Centrifuge the filtrate at 300 x g for 5 min at 4 °C and remove the supernatant. Add 600 µL of HOGMY and disrupt the pellet by pipetting up and down with a 200 µL pipette. Place the tube on ice until the solution becomes ice-cold.
NOTE: If the solution is not ice-cold, the dome will not maintain the proper structure when plated. Avoid introducing bubbles when pipetting ECM-based hydrogel domes, as bubbles will prevent proper adherence to the plate bottom. - Add 600 µL of ice-cold ECM-based hydrogel to suspend the cell pellet and mix well with a 200 µL pipette.
- Remove the 24-well plate from the incubator and place it on a 37 °C plate warmer. Pipette 50 µL of ice-cold HOGMY/ECM-based hydrogel mixture into the center of 24 wells of a 24-well culture plate (50 µL per dome).
- Once plated, let the domes sit on a 37 °C plate warmer for 1 min to allow adherence to the bottom of the plate. Carefully invert the plate, ensure no condensation has accumulated on the bottom of the plate, and place the inverted plate in the incubator at 37 °C and 5% CO2.
- After 15 min, turn the plate right-side-up and pipette 750 µL of room-temperature HOGMY into each well.
NOTE: Pipette media slowly onto the sidewall of wells, being careful not to disrupt the newly formed 3D ECM-based hydrogel dome. - Once all the wells have been filled with media, place the plate in the incubator and change the media every 3 days with 750 µL of HOGM. Passage enteroids every 7-10 days.
- Intestinal enteroid passaging
NOTE: The following protocol is for one 24-well culture plate containing NHP enteroids at a split rate of 1:2.- Remove the enteroid media from each well, being careful not to disrupt the ECM dome containing enteroids, add 500 µL of ice-cold cell dissociation reagent to each well using a 1,000 µL pipette, and incubate at room temperature for 1 min.
- Manually disrupt the ECM by pipetting up and down 8-12x. Transfer the contents of 12 wells to one 15 mL conical tube.
- Once all the solution has been collected from the 24-well culture plate, add 250 µL of cell disruption reagent to each well to ensure all the NHP enteroids have been removed from each well, adding this last rinse to the existing 15 mL conical tubes.
- Place on a rocking platform at 40 rpm for 15 min at room temperature (20 °C). Alternatively, manually shake the tubes for 15 min at room temperature.
- After 15 min, centrifuge at 300 x g for 5 min at 4 °C. Remove the supernatant from the 15 mL tubes until only the pellet remains.
- Add 8 mL of ice-cold DMEM + broad-spectrum antibiotics to one 15 mL conical tube, making sure the pellet is sufficiently disrupted. Centrifuge at 300 x g for 5 min at 4 °C and discard the supernatant.
- Add 600 µL of HOGMY and disrupt the pellet by pipetting up and down with a 1,000 µL pipette. Place the tube on ice until the solution becomes ice-cold. Once the solution is ice-cold, add 600 µL of ice-cold ECM to suspend the cell pellet and mix well with a 1,000 µL pipette. Repeat steps 1.3.2.-1.3.7. for the remaining 12 wells.
- For the remaining steps, refer to steps 1.2.11.-1.2.16.
2. Establishment of enteroid monolayer and scratch wound assay
- Media and treatment preparation
- Chill 5 mL of PBS without calcium or magnesium until ice-cold.
- Prepare DMEM-F12 wash buffer by adding 1 mL of the broad-spectrum antibiotics (100 µg/mL) to 500 mL of DMEM/F12 (Dulbecco's Modified Eagle's Medium/Nutrient Ham's Mixture F12 + 15 mM HEPES buffer). Keep ice-cold.
- Prepare 100 mL of HOGMY, as described in step 1.1.1., and warm it on the benchtop to room temperature.
- Warm 25 mL of 0.25% Trypsin-ethylenediaminetetraacetic acid (EDTA) in a 37 °C water bath.
- Prepare 50 µg/mL, 100 µg/mL, and 200 µg/mL concentrations of HA35 for cell treatment, using sterile PBS without calcium or magnesium as a solvent. The final dilution of HA35 will use HOMGY as the solvent.
NOTE: HA35 preparation may require several dilutions, as HA35 often comes out of solution at a concentration greater than 5 mg/mL.
- Enteroid monolayer formation
NOTE: The following procedure provides volumes sufficient for the generation of one 24-well plate of enteroid monolayers. Adjust the volumes for the desired number of monolayers.- Thaw 96 µL of ECM-based hydrogel BME (phenol red-free) overnight on ice at 2-8 °C. Dilute ECM-based hydrogel in ice-cold PBS without calcium or magnesium at a ratio of 1:50. Coat each well of a 24-well tissue culture plate with 200 µL of ice-cold diluted ECM-based hydrogel.
- Incubate the ECM-based hydrogel-coated plate for a minimum of 1 h at 37 °C and 5% CO2. Aspirate media from the 24-well culture plate containing the 3D enteroids destined for monolayers and replace with 500 µL of cell dissociation reagent/well.
- Manually disrupt ECM-based hydrogel domes by pipetting up and down 8x-12x and transfer the contents of 12 wells to a 15 mL conical tube.
- Wash the 12 empty wells with 250 µL of cell dissociation reagent to ensure all the enteroids have been removed and transfer the contents to a 15 mL conical tube. Repeat step 2.2.5. and step 2.2.6. for the remaining 12 wells using a second 15 mL conical tube.
- Centrifuge the conical tubes at 300 x g for 5 min at 4 °C and discard the supernatant. Add 10 mL of ice-cold DMEM-F12 wash buffer to 15 mL conical tubes and suspend the enteroid pellets by inverting the tubes. Centrifuge at 300 x g for 5 min at 4 °C.
- Remove the supernatant and resuspend the cell pellets in 12 mL of 37 °C Trypsin-EDTA. Place the tubes in a 37 °C water bath for 10 min or until the cells appear soluble. Add 3 mL of DMEM-F12 wash buffer to each tube to dilute Trypsin-EDTA, mix by inverting the tubes, and place on ice.
- Filter the dissociated enteroids through a 37 µM mesh cell strainer, collecting the contents into clean 15 mL conical tubes. Centrifuge the tubes at 300 x g for 5 min at 4 °C.
- While the cells are pelleting in the centrifuge, remove the ECM-based hydrogel-coated plate from the incubator and aspirate any excess ECM-based hydrogel solution without scratching the coated surface or letting the plate completely dry.
- Remove the supernatant from the conical tubes and resuspend the cell pellets with 6 mL of HOMGY. Add 500 µL of the combined 12 mL HOMGY cell suspension into each well of the 24-well plate coated with ECM-based hydrogel at a seeding density of roughly 3 x 105 cells/well. Swirl the plate with a lid on to ensure an even distribution of cells within the wells.
- Incubate the plate at 37 °C and 5% CO2, exchanging HOMGY media every 2 days until the monolayers reach >90% confluency.
- Monolayer treatment and scratch wound assay
- Once the monolayers reach >90% confluency, aspirate media to remove any cells failing to attach. Treat cells with 500 µL of HA35 (50 µg/mL, 100 µg/mL, 200 µg/mL) or PBS, dissolved in HOMGY, for 24 h.
- After 24 h, remove the media, being careful not to disrupt the surface of the monolayer. Using a 200 µL pipette tip, make a linear scratch across the full surface diameter of the monolayer in each well, being careful not to let monolayers dry.
- Wash the scratched monolayers 1x with 500 µL of 1x PBS. Add 500 µL of HA35 (50 µg/mL, 100 µg/mL, 200 µg/mL) or PBS dissolved in HOMGY and transfer the plate to the live-cell analysis instrument (Table of Materials) within a 37 °C and 5% CO2 incubator.
- Under the Schedule icon in the live-cell analysis software (Table of Materials), click on the Launch Add New Vessel Wizard icon and set the scanning frequency to scan on a schedule. Create a new Whole Well vessel under Scan Type, specify the scan settings (i.e., Phase for Image Channels and 4x for Objective), and click on Next.
- Select the plate manufacturer and catalog number from the provided list. Under the Schedule icon, choose the plus symbol where the plate will be placed within the live-cell analysis instrument. Select the desired Scan Pattern, and then create a plate map by selecting "+" on the Plate Layout page. Select Defer Analysis Until Later, and then set the Scanning Schedule for every 4 h for 24 h and verify acquisition settings under the Vessel Wizard Summary page. Finally, select Add to Schedule.
- Once the 24 assay is complete, under the View icon, double-click Vessel Name, and export images and movies. Select wells and a series of images to display and export as a JPEG. Analyze images for percent wound healing using the Wound Healing Size Plugin for ImageJ21, as per step 2.4. below.
- Scratch wound analysis
- Open ImageJ software. Upload the scratch wound assay images (.TIFF or .JPG).
- Select Image > Type > 8-bit to change the image from 24-bit RGB to 8-bit. Install the Wound Healing Size Plugin by selecting Plugins > Macros > Install > Wound Healing Size Plugin.
- Rotate the image so the scratch is vertical and initialize the Wound Healing Size Tool plugin.
- Select the plugin parameters in the dialog box to best fit the scratch wound. Set the value of the parameters for wound healing size options as follows: Variance Window Radius at 20, Threshold value at 100, and Percentage of Saturated Pixels at 0.001, and choose Yes for Set Scale Global. Finalize the selection by clicking on the OK button. Verify if the selected area is the wound area. The above parameters may require standardization from lab to lab.
- Repeat steps 2.4.2.-2.4.4. for the remaining time points for each scratch.
- Calculate the percentage of scratch area wound healing of migrating or proliferating cells over 24 h as follows:
% Wound Healing =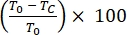
where T0 = % Area at time 0 h and Tc = % Area at 0 h, 4 h, 12 h, or 24 h.
Wyniki
The effects of HA on tissue repair and wound healing in various tissues and organs are well-documented; however, the specific effects of HA with a molecular weight of 35 kDa on fetal or neonatal small intestinal healing and regeneration are currently unknown. To test the ability of HA35 to promote wound healing in a model of the fetal or neonatal small intestine, we generated 3D intestinal enteroids from NHP ileal tissue and further dissociated this tissue into single cells to create 2D enteroid-derived monolayers (
Dyskusje
The gastrointestinal tract of a preterm infant is under continual regenerative pressure from repeated exposures to environmental insults associated with dysbiosis, inflammatory bacterial metabolites and toxins, and intermittent hypoxia23,24. Unfortunately, the intestinal epithelium of the preterm infant is unable to rapidly establish functional integrity23, resulting in barrier dysfunction, increased intestinal permeability, and, in severe...
Ujawnienia
The authors have nothing to disclose and no conflicts of interest.
Podziękowania
This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. HC is supported by grant P20GM134973 from the National Institutes of Health. KB is supported by a Children's Hospital Foundation (CHF) and Presbyterian Health Foundation (PHF) grant. Live-cell imaging services provided by the Cancer Functional Genomics core were supported partly by the National Institute of General Medical Sciences Grant P20GM103639 and National Cancer Institute Grant P30CA225520 of the National Institutes of Health, awarded to the University of Oklahoma Health Sciences Center Stephenson Cancer Center.
Materiały
| Name | Company | Catalog Number | Comments |
| 10 mL Serological Pipet | Fisher Scientific | 13-675-49 | |
| 100x21mm Dish, Nunclon Delta | ThermoFisher Scientific | 172931 | |
| 15 mL Conical tube | VWR | 89039-666 | |
| 24-Well, TC-Treated, Flat Bottom Plate | Corning | 3524 | |
| 37 µM Reversible Cell Strainer | STEMCELL Technologies | 27215 | |
| 50 mL Conical tube | VWR | 89039-658 | |
| 70 µm Sterile Cell Strainers | Fisher Scientific | FB22-363-548 | |
| Albumin, Bovine (BSA) | VWR | 0332-100G | |
| CellTiter-Glo 3D Cell Viability Assay | Promega | G9681 | |
| Dulbecco's Modified Eagle's Medium/Nutrient Ham's Mixture F-12 (DMEM-F12) with 15 mM HEPES buffer | STEMCELL Technologies | 36254 | |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | 100-0485 | |
| ImageJ | NIH | imagej.nih.gov/ij/ | |
| Incucyte S3 Live-Cell Analysis Instrument | Sartorius | 4647 | |
| Incucyte Scratch Wound Analysis Software Module | Sartorius | 9600-0012 | |
| IntestiCult Organoid Growth Medium (Human) | STEMCELL Technologies | 06010 | This is HOGMY, but without the Y-27632 or antibiotics. Also used as base for HOGM, but then only missing the antibiotics. |
| Lipopolysaccharides from Escherichia coli O111:B4, purified by gel filtration chromatography | Millipore Sigma | L3012-10MG | |
| Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenol Red-Free | Corning | 356231 | |
| Nunc MicroWell 96-Well, Nunclon Delta-Treated, Flat-Bottom Microplate | ThermoFisher Scientific | 136101 | |
| PBS (Phosphate-Buffered Saline), 1X [-] Calcium, Magnesium, pH 7.4 | Corning | 21-040-CM | |
| Primocin | Invivogen | ant-pm-1 | This is broad-spectrum antibiotics |
| Sodium Hyaluronate, Research Grade, HA20K | Lifecore Biomedical | HA20K-1 | |
| TC20 Automated Cell Counter | Company: Bio-Rad | 1450102 | |
| Trypsin-EDTA 1X, 0.25% Trypsin | Fisher Scientific | MT25053CI | |
| Y-27632 | STEMCELL Technologies | 72302 |
Odniesienia
- Lemons, J. A., et al. Very low birth weight outcomes of the National Institute of Child Health and Human Development Neonatal Research Network, January 1995 through December 1996. Pediatrics. 107 (1), 1 (2001).
- Burge, K., Vieira, F., Eckert, J., Chaaban, H. Lipid composition, digestion, and absorption differences among neonatal feeding strategies: Potential implications for intestinal inflammation in preterm infants. Nutrients. 13 (2), 550 (2021).
- Duffy, L. C. Interactions mediating bacterial translocation in the immature intestine. The Journal of Nutrition. 130, 432-436 (2000).
- Nanthakumar, N., et al. The mechanism of excessive intestinal inflammation in necrotizing enterocolitis: An immature innate immune response. PLoS One. 6 (3), 17776 (2011).
- Nanthakumar, N. N., Fusunyan, R. D., Sanderson, I., Walker, W. A. Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. Proceedings of the National Academy of Sciences of the United States of America. 97 (11), 6043-6048 (2000).
- He, Y., Lawlor, N. T., Newburg, D. S. Human milk components modulate toll-like receptor-mediated inflammation. Advances in Nutrition. 7 (1), 102-111 (2016).
- Walker, W. A., Iyengar, R. S. Breast milk, microbiota, and intestinal immune homeostasis. Pediatric Research. 77 (1-2), 220-228 (2015).
- Westerbeek, E. A., vanden Berg, A., Lafeber, H. N., Fetter, W. P., van Elburg, R. M. The effect of enteral supplementation of a prebiotic mixture of non-human milk galacto-, fructo- and acidic oligosaccharides on intestinal permeability in preterm infants. British Journal of Nutrition. 105 (2), 268-274 (2011).
- vanden Berg, A., et al. The effect of glutamine-enriched enteral nutrition on intestinal permeability in very-low-birth-weight infants: A randomized controlled trial. Journal of Parenteral and Enteral Nutrition. 30 (5), 408-414 (2006).
- Foster, J. P., Seth, R., Cole, M. J. Oral immunoglobulin for preventing necrotizing enterocolitis in preterm and low birth weight neonates. The Cochrane Database of Systematic Reviews. 4 (4), (2016).
- Maffei, D., Schanler, R. J. Human milk is the feeding strategy to prevent necrotizing enterocolitis. Seminars in Perinatology. 41 (1), 36-40 (2017).
- Patel, A. L., Kim, J. H. Human milk and necrotizing enterocolitis. Seminars in Pediatric Surgery. 27 (1), 34-38 (2018).
- Nolan, L. S., Parks, O. B., Good, M. A review of the immunomodulating components of maternal breast milk and protection against necrotizing enterocolitis. Nutrients. 12 (1), 14 (2019).
- Hill, D. R., et al. Human milk hyaluronan enhances innate defense of the intestinal epithelium. Journal of Biological Chemistry. 288 (40), 29090-29104 (2013).
- Burge, K., Bergner, E., Gunasekaran, A., Eckert, J., Chaaban, H. The role of glycosaminoglycans in protection from neonatal necrotizing enterocolitis: A narrative review. Nutrients. 12 (2), 546 (2020).
- Chaaban, H., et al. Acceleration of small intestine development and remodeling of the microbiome following hyaluronan 35 kDa treatment in neonatal mice. Nutrients. 13 (6), 2030 (2021).
- Gunasekaran, A., et al. Hyaluronan 35 kDa enhances epithelial barrier function and protects against the development of murine necrotizing enterocolitis. Pediatric Research. 87 (7), 1177-1184 (2020).
- Montenegro-Miranda, P. S., et al. A novel organoid model of damage and repair identifies HNF4α as a critical regulator of intestinal epithelial regeneration. Cellular and Molecular Gastroenterology and Hepatology. 10 (2), 209-223 (2020).
- Lee, C., Hong, S. N., Kim, E. R., Chang, D. K., Kim, Y. H. Epithelial regeneration ability of Crohn's disease assessed using patient-derived intestinal organoids. International Journal of Molecular Sciences. 22 (11), 6013 (2021).
- Gurung, S., et al. Maternal Zika virus (ZIKV) infection following vaginal inoculation with ZIKV-infected semen in timed-pregnant olive baboons. Journal of Virology. 94 (11), 00058 (2020).
- Suarez-Arnedo, A., et al. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS One. 15 (7), 0232565 (2020).
- Kobelt, D., Walther, W., Stein, U. S. Real-time cell migration monitoring to analyze drug synergism in the scratch assay using the IncuCyte system. Methods in Molecular Biology. 2294, 133-142 (2021).
- de Jong, J. C. W., Ijssennagger, N., van Mil, S. W. C. Breast milk nutrients driving intestinal epithelial layer maturation via Wnt and Notch signaling: Implications for necrotizing enterocolitis. Biochimica et Biophysica Acta - Molecular Basis of Disease. 1867 (11), 166229 (2021).
- Yu, Y., et al. Erythropoietin protects epithelial cells from excessive autophagy and apoptosis in experimental neonatal necrotizing enterocolitis. PLoS One. 8 (7), 69620 (2013).
- Kessler, S. P., et al. Multifunctional role of 35 kilodalton hyaluronan in promoting defense of the intestinal epithelium. The Journal of Histochemistry and Cytochemistry. 66 (4), 273-287 (2018).
- Fraser, J. R. E., Laurent, T. C., Laurent, U. B. G. Hyaluronan: Its nature, distribution, functions and turnover. Journal of Internal Medicine. 242 (1), 27-33 (1997).
- Kim, Y., et al. Hyaluronan 35kDa treatment protects mice from Citrobacter rodentium infection and induces epithelial tight junction protein ZO-1 in vivo. Matrix Biology. 62, 28-39 (2017).
- Prehm, P., Schumacher, U. Inhibition of hyaluronan export from human fibroblasts by inhibitors of multidrug resistance transporters. Biochemical Pharmacology. 68 (7), 1401-1410 (2004).
- Stenson, W. F., Ciorba, M. A. Nonmicrobial activation of TLRs controls intestinal growth, wound repair, and radioprotection. Frontiers in Immunology. 11 (3591), 617510 (2021).
- Riehl, T. E., Ee, X., Stenson, W. F. Hyaluronic acid regulates normal intestinal and colonic growth in mice. American Journal of Physiology-Gastrointestinal and Liver Physiology. 303 (3), 377-388 (2012).
- Riehl, T. E., Santhanam, S., Foster, L., Ciorba, M., Stenson, W. F. CD44 and TLR4 mediate hyaluronic acid regulation of Lgr5+ stem cell proliferation, crypt fission, and intestinal growth in postnatal and adult mice. American Journal of Physiology-Gastrointestinal and Liver Physiology. 309 (11), 874-887 (2015).
- Hill, D. R., Kessler, S. P., Rho, H. K., Cowman, M. K., de la Motte, C. A. Specific-sized hyaluronan fragments promote expression of human beta-defensin 2 in intestinal epithelium. Journal of Biological Chemistry. 287 (36), 30610-30624 (2012).
- Fernando, E. H., Gordon, M. H., Beck, P. L., MacNaughton, W. K. Inhibition of intestinal epithelial wound healing through protease-activated receptor-2 activation in Caco2 cells. Journal of Pharmacology and Experimental Therapeutics. 367 (2), 382-392 (2018).
- Nyegaard, S., Christensen, B., Rasmussen, J. T. An optimized method for accurate quantification of cell migration using human small intestine cells. Metabolic Engineering Communications. 3, 76-83 (2016).
- Roodsant, T., et al. A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Frontiers in Cellular and Infection Microbiology. 10, 272 (2020).
- Singh, A., Poling, H. M., Spence, J. R., Wells, J. M., Helmrath, M. A. Gastrointestinal organoids: a next-generation tool for modeling human development. American Journal of Physiology-Gastrointestinal and Liver Physiology. 319 (3), 375-381 (2020).
- Foulke-Abel, J., et al. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Experimental Biology and Medicine. 239 (9), 1124-1134 (2014).
- Foulke-Abel, J., et al. Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology. 150 (3), 638-649 (2016).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone