A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Performing Custom MicroRNA Microarray Experiments
In This Article
Summary
A simple procedure of performing custom microRNA microarray experiments is described. The steps include isolating RNA, labeling RNA and reference DNA, hybridizing the samples to microarrays, scanning the microarrays, quantifying and analyzing hybridization signals.
Abstract
microRNAs (miRNAs) are a large family of ˜ 22 nucleotides (nt) long RNA molecules that are widely expressed in eukaryotes 1. Complex genomes encode at least hundreds of miRNAs, which primarily inhibit the expression of a vast number of target genes post-transcriptionally 2, 3. miRNAs control a broad range of biological processes 1. In addition, altered miRNA expression has been associated with human diseases such as cancers, and miRNAs may serve as biomarkers for diseases and prognosis 4, 5. It is important, therefore, to understand the expression and functions of miRNAs under many different conditions.
Three major approaches have been employed to profile miRNA expression: real-time PCR, microarray, and deep sequencing. The technique of miRNA microarray has the advantage of being high-throughput, generally less expensive, and most of the experimental and analysis steps can be carried out in a molecular biology laboratory at most universities, medical schools and associated hospitals. Here, we describe a method for performing custom miRNA microarray experiments. A miRNA probe set will be printed on glass slides to produce miRNA microarrays. RNA is isolated using a method or reagent that preserves small RNA species, and then labeled with a fluorescence dye. As a control, reference DNA oligonucleotides corresponding to a subset of miRNAs are also labeled with a different fluorescence dye. The reference DNA will serve to demonstrate the quality of the slide and hybridization and will also be used for data normalization. The RNA and DNA are mixed and hybridized to a microarray slide containing probes for most of the miRNAs in the database. After washing, the slide is scanned to obtain images, and intensities of the individual spots quantified. These raw signals will be further processed and analyzed as the expression data of the corresponding miRNAs. Microarray slides can be stripped and regenerated to reduce the cost of microarrays and to enhance the consistency of microarray experiments. The same principles and procedures are applicable to other types of custom microarray experiments.
Protocol
1. Printing of custom miRNA microarrays
- Print the microarray slides using the microarray core facility services at a university or a company. The quality of microarray fabrication is one of the most critical factors for the success of a microarray experiment. Try a few services if possible. Ideally, one would print 50-100 slides at a time, and all the slides will look identical, with well separated, individual spots.
- For microarray support, we use the GAPS II coated slides.
- For miRNA probes, we use the NCode Multi-Species miRNA Microarray Probe Set V2.
- Dissolve all the oligonucleotides in 3 x SSC at 10 μM and quadruply print them on the slides. Fix the slides by ultraviolet irradiation according to instructions for the GAAPS II slides. Label the slide with a diamond pen to mark the area and side with the printed probes. Store at 22°C.
2. Sample preparation
- Any method or reagent can be used to isolate RNA, as long as it preserves small RNAs. We generally use Trizol (Invitrogen) to extract total RNA, with satisfactory quantity and quality of RNA preparations, as measured by A260nm and A280nm readings.
- For RNA labeling, mix ˜ 25 μg of total RNA with 0.5 μg of 5'-pCU-DY547-3' 6 in 1 x buffer (50 mM HEPES, pH 7.8, 20 mM MgCl2, 10 μg/ml BSA, 10% DMSO) containing ˜ 20 units T4 RNA Ligase 1, supplemented with ˜ 10 mM DTT and ˜ 0.02-0.03 final volume of the 10 x T4 RNA Ligase 1 buffer for ATP.
- Allow labeling to proceed on ice inside a refrigerator for 2-24 hours. Precipitate RNA with 0.3 M sodium acetate and 3 volumes of ethanol. For efficient ligation and precipitation, dissolve total RNA at over 2 mg/ml, and keep the total ligation volume at ˜ 20 μl.
- If less RNA is available, the amount of input RNA and 5'-pCU-DY547-3' can be scaled down.
- If the RNA is very dilute, add carrier such as yeast tRNA to aid in precipitation.
- Combine and label in total 1 μg of reference DNA oligonucleotides corresponding to a subset of mammalian miRNAs using the Ulysis Alexa Fluor 647 Nucleic Acid Labeling Kit 6 as a control for the hybridization process. Purify the labeled DNA using a CentriSep column and save in dark at -20°C.
3. Microarray hybridization
- To use the slides for the first time, pre-hybridize the slides in a filtered solution of 3 x SSC, 0.1% SDS, 0.2% BSA for 30-60 min at 37 °C. Submerge them in water a few times, then in isopropanol. Dry the slides using a centrifuge with a slide adapter at 100 g for 5 min at 22°C.
Note: We use Corning microarray hybridization chambers and Erie Scientific's mSeries of Lifterslips to perform microarray hybridization.
- Rinse the lifterslips in water, then in ethanol, before drying in air. Place a slide inside a microarray hybridization chamber base, and a lifterslip on top of the slide. The lifterslip will cover the probe area, with its white strips contacting the slide.
- In the dark, spin down the precipitated, labeled RNA. Wash it once with 70% ethanol, and air dry the pellet. The pellet should be reddish.
- Prepare a hybridization solution containing 400 mM Na2HPO4 pH 7.0, 0.8% BSA, 5% SDS, 12% formamide 6 with 1/15-1/100 μl of the purified, labeled reference DNA (2.4) per RNA sample. The amount of added reference DNA depends on how many different oligonucleotides are labeled (2.4). If there are over 100, then less dilution is needed.
- Dissolve the RNA pellet well with ˜ 60 μl of the hybridization solution.
- Add 20 μl of water to each of the humidifying wells in the Corning microarray hybridization chamber base.
- Add the mixture of labeled RNA and reference DNA to the slide. Use a thin pipet tip, gently touch the edge of the lifterslip, and allow the solution to enter the space between the lifterslip and slide through suction.
- Place the hybridization chamber cover over the base, then seal the chamber with metal clips.
- Put the whole chamber inside the plastic bag that comes with the chamber cassette from Corning.
- Place the chamber cassette(s) inside a container with wetted paper towel; cover the container with plastic wrap and place it inside a 37°C humid, cell culture incubator for ˜ 24 hours. These precautions (3.6, 3.9, 3.10) ensure that the hybridization solution does not dry out during incubation.
4. Post-hybridization processing
- Disassemble the chamber cassettes one by one. Submerge a slide and its lifterslip in 2 x SSC at 22°C. The lifterslip will naturally fall off the slide. Place it in 0.8 x SSC. Wash the slides in 0.8 x SSC twice, then three times in 0.4 x SSC, 1-2 min in total. Dry the slides using a centrifuge with a slide adapter. Wash the lifterslips with water and save for re-use later.
- Scan the slides with any suitable scanner, e.g., the Perkin Elmer ScanArray 5000 or Molecular Devices Axon GenePix 4000B microarray scanner. Scan at wavelengths close to 547 nm and 647 nm to obtain the corresponding image files.
- Use programs such as BlueFuse to quantify pixel intensities in the input, image files from 4.2). Inspect individual spots with the program to exclude abnormal spots on the microarrays from further consideration. These abnormal spots usually arise from poor slide printing or slide handling after hybridization.
- Use programs such as GeneSpring and Excel for data analysis and presentation. General considerations for microarray data normalization apply, which is beyond the scope of this paper.
- Once satisfactory signals are obtained after 4.3), strip the microarray slides for reuse 7. Rinse the slides in water, then immerse in pre-warmed, 1 mM NaOH and 0.1 x SSC in a staining dish at 62°C for 10-20 minutes. Repeat the incubation once. Wash the slides a few times in water for up to 60 minutes with gentle shaking at 22°C. Dry the slides using a centrifuge, and store at 22°C.
- To use the regenerated slides, it is not necessary to pre-hybridize them again. Simply wash them in water followed by isopropanol, and dry the slides right before hybridization.
5. Representative Results:
We have largely followed the described procedures to profile global miRNA expression in thousands of samples, i.e., RNAs isolated from zebrafish to human specimen under many different conditions. Figure 1 shows a scanned image of a microarray to demonstrate very precise and strong hybridization signals on the slide. The Pearson correlation coefficients between technical replicates of microarray hybridization are ˜ 0.99 7, indicating excellent reproducibility.
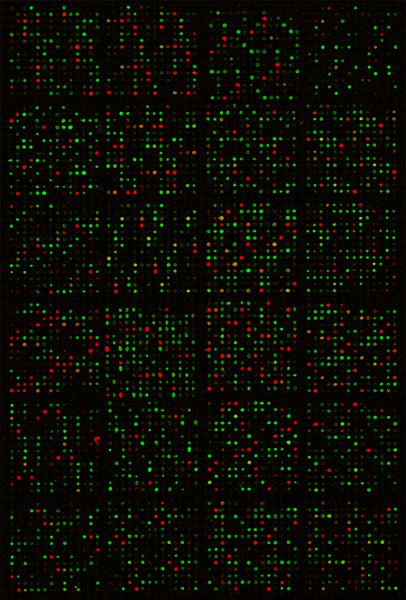
Figure 1 Composite image of a scanned miRNA microarray slide after hybridization. Red spots resulted from hybridizations by the reference DNA, green spots from the DY547-labelled RNA sample, while the yellow spots were from hybridizations by both the DNA and RNA to the same probes.
Discussion
Despite recent advances in deep sequencing technologies, microarray remains a viable choice for high-throughput analysis of DNA and RNA. Compared to deep sequencing, microarray experiments are cheaper, and a typical molecular biology laboratory can perform most of the experiments and data analysis in-house, which allows for flexibility and saves time. In the future, microarrays are likely well-suited to intensively interrogate sets of genes, e.g., all or a subset of the transcription factors in a genome or miRNAs, and ...
Disclosures
No conflicts of interest declared.
Acknowledgements
The work was supported in part by National Institute of Drug Abuse Center (P50 DA 011806) and United States Army Department of Defense (W81XWH-07-1-0183).
Materials
| Name | Company | Catalog Number | Comments |
| NCode Multi-Species miRNA Microarray Probe Set V2 | Invitrogen | MIRMPS201 | Designed based on the miRBase Release 9.0 (October 2006). It contains ˜ 1,140 unmodified, 34-44 nt long oligonucleotides as probes for worm, fly, zebrafish, mouse, rat, and human miRNAs, and a number of internal control probes such as snoRNAs. The miRNA probes are doublets of the sequences complementary to mature miRNAs, hence the size of ˜ 44 nt. For analysis one can focus on miRNAs from a particular genome(s) of interest. |
| Trizol | Invitrogen | 15596018 | We have also used enriched, small RNA fraction for labeling, although total RNA samples are faster and easier to prepare and to quantify and suitable for downstream applications such as mRNA analysis. |
| T4 RNA Ligase 1 | New England Biolabs | M0204L | |
| Ulysis Alexa Fluor 647 Nucleic Acid Labeling Kit | Invitrogen | U21660 | This kit or similar products can be used to label experimental RNA samples or a control RNA (instead of control DNA) as well. |
| 5’-pCU-DY547-3’ | Dharmacon | Custom made | Small RNA fraction can be similarly labeled by ligation. |
| CentriSep columns | Princeton Separations | CS-901 | |
| GAPSII coated slide | Corning | 40004 | Other types of slides may be also used. |
| Microarray hybridization chambers | Corning | 2551 or 40080 | Other kinds of hybridization chambers and coverslips should also work. Using commercially available hybridization machines can reduce hybridization time significantly, e.g., to ˜ 2 hours. |
| Lifterslips | Thermo Fisher Scientific, Inc. | 25X60I-M5439-001-LS | |
| BlueFuse | BlueGenome | ||
| GeneSpring | Agilent Technologies |
References
- Ambros, V. The functions of animal microRNAs. Nature. 431, 350-355 (2004).
- Friedman, R. C., Farh, K. K., Burge, C. B., Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92-105 (2009).
- Chekulaeva, M., Filipowicz, W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21, 452-460 (2009).
- Farazi, T. A., Spitzer, J. I., Morozov, P., Tuschl, T. miRNAs in human cancer. J. Pathol. 223, 102-115 (2011).
- Small, E. M., Olson, E. N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 469, 336-342 (2011).
- Thomson, J. M., Parker, J., Perou, C. M., Hammond, S. M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 1, 47-53 (2004).
- Zhang, X., Xu, W., Tan, J., Zeng, Y. Stripping custom microRNA microarrays and the lessons learned about probe:slide interactions. Anal. Biochem. 386, 222-227 (2009).
- Griffiths-Jones, S., Saini, H. K., van Dongen, S., Enright, A. J. miRBase: tools for microRNA genomics. Nucl. Acids. Res. 36, D154-D158 (2008).
- Landgraf, P. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129, 1401-1414 (2007).
- Chiang, H. R. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes. Dev. 24, 992-1009 (2010).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved