A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
An ELISA Based Binding and Competition Method to Rapidly Determine Ligand-receptor Interactions
In This Article
Summary
The presented protocols describe two enzyme-linked immunosorbent assay (ELISA) based techniques for the rapid investigation of ligand-receptor interactions: The first assay allows the determination of dissociation constant between ligand and receptor. The second assay enables a rapid screening of blocking peptides for ligand-receptor interactions.
Abstract
A comprehensive understanding of signaling pathways requires detailed knowledge regarding ligand-receptor interaction. This article describes two fast and reliable point-by-point protocols of enzyme-linked immunosorbent assays (ELISAs) for the investigation of ligand-receptor interactions: the direct ligand-receptor interaction assay (LRA) and the competition LRA. As a case study, the ELISA based analysis of the interaction between different lambda interferons (IFNLs) and the alpha subunit of their receptor (IL28RA) is presented: the direct LRA is used for the determination of dissociation constants (KD values) between receptor and IFN ligands, and the competition LRA for the determination of the inhibitory capacity of an oligopeptide, which was designed to compete with the IFNLs at their receptor binding site. Analytical steps to estimate KD and half maximal inhibitory concentration (IC50) values are described. Finally, the discussion highlights advantages and disadvantages of the presented method and how the results enable a better molecular understanding of ligand-receptor interactions.
Introduction
A comprehensive understanding of signaling pathways requires detailed knowledge about the ligand-receptor interaction. Most methods for assessing the interaction of a particular ligand with its specific receptor are expensive, time consuming, labor intensive and require specific equipment and expertise 1.
This article describes two fast and reliable point-by-point protocols to investigate the ligand-receptor interaction based on an enzyme linked immunosorbent assay (ELISA): the direct ligand-receptor interaction assay (LRA) and the competition LRA. ELISA is a highly sensitive, specific and readily available technique, routinely used in almost every laboratory. ELISA can be performed and adapted in various fashions. The presented protocols are optimized for the investigation of the interaction between different lambda interferons (INFLs) and their receptor.
The direct LRA allows for a quantification of ligand-receptor binding with respect to ligand concentration and thus yields a binding curve. Using an appropriate model for the ligand-receptor interaction, the data can be further analyzed to estimate the dissociation constant (KD).
In the presented protocol, the commonly used Hill equation is applied to model the ligand-receptor binding. Although other methods such as the surface plasmon resonance technology 2,3 allow the determination of the binding affinities between two proteins, this technology is often labor intensive, expensive, and requires special laboratory equipment.
The competition LRA enables the screening of inhibitory peptides: The ligand-receptor binding is quantified with respect to peptide concentration. This yields a dose-response curve describing the inhibitory effect of the peptide. The data can be further analyzed to estimate the half maximal inhibitory concentration (IC50) of the blocking peptide.
Both ELISA protocols are easy to use and can be adapted to a broad range of research questions. Recombinant proteins of any kind can be used to reliably and fast determine the interaction parts. In addition, the competition LRA can be used to determine critical interaction sites of ligands and receptors by using blocking peptides, which are designed to mimic either the ligand or the receptor. If the blocking peptide shows efficient and specific inhibition, the peptide occupies a critical interaction site of the ligand (if the peptide mimics the receptor) or of the ligand (if the peptide mimics the ligand).
The first protocol describes the KD value determination of different INFLs and the alpha subunit of their receptor, i.e., the interleukin-28 receptor (IL28RA) using the direct LRA. Next, the second protocol shows how to determine the capability of a 20 amino acid long peptide to inhibit the INFL-IL28RA interactions. The peptide is designed to compete with IFNLs at their receptor binding site and thus enables a molecular understanding of the interaction. Furthermore, this peptide can be used to block IL28RA in in vitro experiments to determine the impact on downstream signaling effects4.
Protocol
1. Reagent Preparation
- To prepare carbonate coating buffer, dissolve 0.36 g Na2CO3 and 0.84 g NaHCO3in 100 ml distilled water; sterile filter the buffer by using a vacuum driven 0.22 µm polyethersulfone (PES) membrane filter and store at RT until usage.
- Prepare washing solution by adding 0.05% v/v Tween 20 in Phosphate buffered saline (PBS).
- Prepare a 5% Bovine Serum Albumin (BSA) (blocking solution) in PBS solution by dissolving 5 g BSA (≥98%) in 100 ml PBS and store at 4 °C.
- Recombinant Receptor, Ligands and Blocking Peptides
- Reconstitutethe recombinant human interleukin receptor alpha subunit (IL28RA) and recombinant His-tagged ligands of human IFN (IFNL1-3) according to the manufacturer's instructions and store at -80°C. Synthetize blocking peptides and used as previously described 4. Use PBS to prepare different concentrations of ligands and peptides for use in the assays.
- To prepare the primary antibody, dilute 6x His Mouse monoclonal antibody in PBS with 0.1% BSA at 1:1,000 dilution. To prepare the secondary antibody, dilute horseradish peroxidase (HRP) conjugated goat anti-mouse IgG (H+L) in PBS with 0.1% BSA at 1:10,000 dilution.
- Prepare TMB solution by mixing the reagents A and B according to the manufacturer's instructions.
- Prepare stop solution by adding 5 N sulphuric acid (H2SO4) in distilled water and store at RT.
2. Enzyme-linked Immunosorbent Assays (ELISAs)
NOTE: The direct ligand-receptor interaction ELISA (direct LRA, Figure 1) can be used to measure the receptor-ligand dissociation constant (KD), as a measure of the receptor-ligand binding affinity. The competition ligand-receptor interaction ELISA (competition LRA, Figure 2) allows screening of peptides (and other blocking compounds), which act to interfere with the interaction between ligand and receptor. The basic protocol that was previously published 5 was further optimized.
NOTE: In both ELISA methods use multichannel pipette for adding solutions to the wells of 96-well plate in each step. In solution decant or washing steps, throw out the solutions directly into the sink.
- Direct Ligand-Receptor-Interaction Assay (direct LRA)
NOTE: For an illustration of the workflow (see Figure 1).- Coating Plate with Recombinant Receptor
- Dilute the recombinant receptor in carbonate buffer to a final concentration of 100 ng/µl. Coat wells of 96-well microtitre plate with fixed receptor concentration (100 ng/µl) by pipetting 100 µl to each well using a multichannel pipette. Exclude outer walls of the plate to avoid well edge artifact. Cover the plate with a lid and incubate the plate at 4 °C O/N.
- Blocking and Addition of Ligands
- The next day, remove the coating solution by tilting the plate against the sink and wash the plate 3 times with washing solution (PBS + 0.05% v/v Tween 20).
- Block the free receptor-binding sites in the coated plate using 200 µl of 5% BSA solution to each well using a multichannel pipette and incubate the plate for 2 hr at RT.
- Discard the blocking solution (see step 2.1.2.1.) and wash the plate 3 times with washing solution.
- Prepare the recombinant His-tagged ligands at different concentrations (e.g., 8 µg/ml, 4 µg/ml, 2 µg/ml, 1 µg/ml, 0.5 µg/ml, 0.25 µg/ml, 0.125 µg/ml, 0.063 µg/ml, 0.031 µg/ml, 0.0 µg/ml) in PBS. Add only PBS in the blank wells.
- Add 100 µl of each ligand concentration to the wells in duplicate and incubate the plate for 2 hr at RT allowing receptor-ligand interaction.
- Incubation with Antibodies
- Following incubation with the ligands, wash the plate 3 times with washing solution.
- Pipette 100 µl of primary anti-His mouse monoclonal antibody solution (1:1,000) to each well.
- Incubate the plate at RT for 2 hr; after incubation, discard the antibody solution (see step 2.1.2.1.) and wash the plate 3 times with washing solution.
- Add 100 µl of HRP coupled goat anti-mouse IgG secondary antibody solution (1:10,000) to each well. Incubate the plate for 45 min at RT.
- Discard the antibody solution (see step 2.1.2.1.) and wash the plate 3 times with washing solution.
- Addition of Substrate and Development
- Bring the TMB substrate solutions to RT and prepare TMB substrate solution A and B at 1:1 ratio. Add 100 µl freshly prepared substrate to each well and keep the plate at RT for 15-30 min. After sufficient color development add 50 µl stop solution.
- Reading the Plate and Data Analysis
NOTE: The described protocol is based on the assumption that the measured signal rises from specific binding. It might be necessary to estimate the contribution of unspecific binding to the signal but this is out of the scope of this protocol.- Read the absorbance (optical density, OD) directly at 450 nm.
- Subtract the background signal from the measured OD values and normalize them. Transform all values of the ligand concentration to logarithmic scale (base 10, log10).
- Plot the normalized and background corrected OD values (Y-axis, corresponds to the fraction of occupied receptor binding sites) against the logarithm of the ligand concentration (X-axis, log10 scale).
- To estimate the KD value, fit the data to the following form of the Hill equation:
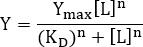
NOTE: Here Y denotes the fraction of occupied receptor binding sites and Ymax the maximal binding; [L] denotes the concentration of free ligand and the Hill coefficient. If there is only one binding site for the ligand, the Hill coefficient is n = 1. For systems with more than one ligand binding site, the binding exhibits positive cooperativity if n >1, negative cooperativity if n<1 and no cooperativity if n = 1. The microscopic dissociation constant is termed and corresponds to the half maximal effective concentration EC506. The apparent dissociation constant is Kd = (KD)n. In the simplest case where n = 1, the dissociation constant corresponds to the ligand concentration at which half of the receptor binding sites are occupied and Kd = KD. This model assumes mass action binding under equilibrium conditions, as well as that only a small fraction of the added ligand is bound to the receptor, i.e., [L] >> [RL].
- Coating Plate with Recombinant Receptor
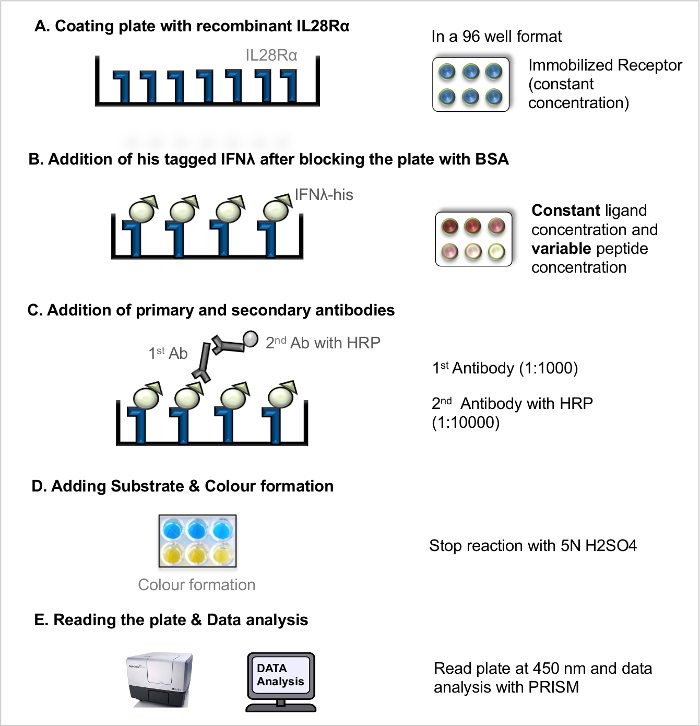
Figure 1. Direct ligand-receptor-interaction assay (direct LRA). Step-by-step protocol for direct LRA. Please click here to view a larger version of this figure.
- Competition Ligand-Receptor-Interaction Assay (competition LRA)
NOTE: For an illustration of the workflow see Figure 2. The competition LRA procedure follows the same steps as the direct LRA (coating the plate, antibody incubation, plate development) except for important changes in the ligand and peptides addition step. Proper negative controls are essential for this assay. In a previous screening study 4, the scrambled blocking peptide did not show antagonistic effects.- Blocking - Addition of Ligands and Blocking Peptides
- The next day, remove the coating solution and wash the plate (see 2.1.2.1).
- Block the coated plate by adding 200 µl of 5% BSA solution to each well and incubate the plate for 2 hr at RT.
- Prepare the recombinant His-tagged ligands (IFNL1-3) at a fixed concentration (2x-20 ng/ml) in PBS.
- Prepare the blocking peptide (cf. Table 3) with different concentrations ranging from 10 nM to 100 µM in PBS to guarantee a dose-response curve.
NOTE: This enables subsequent determination of the IC50 value for the blocking peptide. In control wells, add only fixed ligand concentration without peptide to derive the maximum (100%) binding. In the blank, add only PBS without ligand or peptide. - Add 50 µl of the ligands (IFNL1-3) and 50 µl of each peptide concentration to the wells in duplicates.
- Incubate the plate for 2 hr at RT.
- Reading the Plate and data Analysis
NOTE: The described protocol is based on the assumption that the measured signal rises from specific binding. It might be necessary to estimate the contribution of unspecific binding to the signal but this is out of the scope of this protocol.- Read the absorbance (optical density, OD) directly at 450 nm.
- Subtract the background signal from the measured OD values and normalize them. Transform all values of the peptide concentration to logarithmic scale (base 10, log10).
- Plot the normalized and background corrected OD values (Y-axis, corresponds to the fraction of occupied receptor binding sites) against the logarithm of the ligand concentration (X-axis, log10 scale).
- To estimate the IC50 value, fit the data to the following equation:
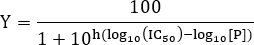
NOTE: Here [P] is the peptide concentration and the Hill slope. The Hill slope describes the steepness of the dose-response curve. The IC50 corresponds to the inhibitor concentration at which 50% inhibition of binding between ligand and receptor is observed.
- Blocking - Addition of Ligands and Blocking Peptides

Figure 2. Competition ligand-receptor-interaction assay (competition LRA). Step-by-step protocol for competition LRA. Please click here to view a larger version of this figure.
Results
The dissociation constants between INFL1-3 and their receptor alpha subunit IL28RA were determined using the direct LRA. The results are shown in Figure 3: The fraction of occupied binding sites is plotted against the logarithm of the respective IFN concentration. The Scatchard plot of the data is shown in the bottom right corner. The results illustrate that the direct LRA yields a binding curve, which can be further analyzed to estimate the KD value. The K
Discussion
ELISA is a standard and well-established method for many laboratories. We have further modified and improved a previously published method 5,7. The demonstrated step-by-step protocol shows how it can be used in a simple way to determine the KD values of ligand-receptor interactions. In addition, the IC50 of a blocking peptide that interferes with the ligand-receptor interaction can be determined.
Major advantages are the rapid setup, easy preparation of reagents and famil...
Disclosures
A.E. was supported by a research grants from the "SNSF Ambizione Score" program (PZ00P3_154709), "Forschungsfond, Förderung strategischer Projekte" University of Basel, Stiftungsinfektionskrankheiten Basel, and Bangeter Rhyner Stiftung. J.L. acknowledges support by an iPhD fellowship of the SystemsX.ch initiative in systems biology program (9th call).
Acknowledgements
We thank Prof. J. Stelling (Department of Biosystems Science and Engineering, ETH Zurich and Swiss Institute for Bioinformatics, Basel, Switzerland) for his critical review of the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| Nunc-Immunoplate (F96 Maxi sorp) | Thermo Scientific | 442404 | ELISA plate |
| Sodium carbonate (Na2CO3) | Merck | 497-19-8 | For ELISA plate coating buffer |
| Sodium hydrogen carbomnate(NaHCO3) | Merck | 144-55-8 | For ELISA plate coating buffer |
| Bovine Serum Albumin (BSA) | Sigma | A7030-100G | 5% BSA in PBS for Blocking |
| rhIL-28Rα/IFNλR1 | R&D systems | 5260-MR | Recombinant human interlukin-28 Receptor alpha |
| rhIL-29/IFNλ1 | R&D systems | 1598-IL/CF | Recombinant human interlukin-29/Carrier free/C-terminal 10-His tag |
| rhIL-28A/IFNλ2 | R&D systems | 1587-IL/CF | Recombinant human interlukin-28A/Carrier free/C-terminal 6-His tag |
| rhIL-28B/IFNλ3 | R&D systems | 5259-IL/CF | Recombinant human interlukin-28B/Carrier free/C-terminal 6-His tag |
| 6x His Monoclonal antibody (Mouse) | Clontech | 631212 | Primary antiboy to capture His tagged Ligands |
| Goat anti-Mouse igG (H+L) | Jackson Immuno Research | 115-035-166 | Horseradish Peroxidase conjucated secondary antibody |
| BDoptEIA TMB reagent set | BD Biosciences | 555214 | ELISA - TMB substrate solution |
| Sulfuric acid (H2SO4) | Fulka | 84720 | 5 N H2SO4 (Enzyme reaction stop solution) |
| Synergy/H1 - Microplate reader | BioTeK | ELISA plate reader |
References
- Schneider, P., Willen, L., Smulski, C. R. Tools and techniques to study ligand-receptor interactions and receptor activation by TNF superfamily members. Methods in enzymology. 545, 103-125 (2014).
- Rossi, G., et al. Biosensor analysis of anti-citrullinated protein/peptide antibody affinity. Analytical biochemistry. 465, 96-101 (2014).
- van der Merwe, P. A., Barclay, A. N. Analysis of cell-adhesion molecule interactions using surface plasmon resonance. Curr Opin Immunol. 8, 257-261 (1996).
- Egli, A., et al. IL-28B is a key regulator of B- and T-cell vaccine responses against influenza. PLoS Pathog. 10, e1004556 (2014).
- Rosenbluh, J., et al. Positively charged peptides can interact with each other, as revealed by solid phase binding assays. Analytical biochemistry. 352, 157-168 (2006).
- Goutelle, S., et al. The Hill equation: a review of its capabilities in pharmacological modelling. Fundamental & clinical pharmacology. 22, 633-648 (2008).
- Levin, A., et al. Peptides derived from HIV-1 integrase that bind Rev stimulate viral genome integration. PLoS One. 4, e4155 (2009).
- Egli, A., Santer, M. D., O'Shea, D., Tyrrell, D. L., Houghton, M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerging infectious diseases. , e51 (2014).
- Gad, H. H., Hamming, O. J., Hartmann, R. The structure of human interferon lambda and what it has taught us. J Interferon Cytokine Res. 30, 565-571 (2010).
- Folch, B., Rooman, M., Dehouck, Y. Thermostability of salt bridges versus hydrophobic interactions in proteins probed by statistical potentials. Journal of chemical information and modeling. 48, 119-127 (2008).
- Yuzlenko, O., Lazaridis, T. Interactions between ionizable amino acid side chains at a lipid bilayer-water interface. The journal of physical chemistry. B. 115, 13674-13684 (2011).
- Tissot, A. C., Vuilleumier, S., Fersht, A. R. Importance of two buried salt bridges in the stability and folding pathway of barnase. Biochemistry. 35, 6786-6794 (1996).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved