A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
A Novel In Vitro Wound Healing Assay to Evaluate Cell Migration
In This Article
Summary
Here, we present a protocol to evaluate the effect of peptides on the migration of bronchial epithelial cells. This method allows for the rapid and highly reproducible obtainment of quantitative data on the speed of cell migration and wound closure.
Abstract
The aim of this work is to show a novel method to evaluate the ability of some immunomodulatory molecules, such as antimicrobial peptides (AMPs), to stimulate cell migration. Importantly, cell migration is a rate-limiting event during the wound-healing process to re-establish the integrity and normal function of tissue layers after injury. The advantage of this method over the classical assay, which is based on a manually made scratch in a cell monolayer, is the usage of special silicone culture inserts providing two compartments to create a cell-free pseudo-wound field with a well-defined width (500 μm). In addition, due to an automated image analysis platform, it is possible to rapidly obtain quantitative data on the speed of wound closure and cell migration. More precisely, the effect of two frog-skin AMPs on the migration of bronchial epithelial cells will be shown. Furthermore, pretreatment of these cells with specific inhibitors will provide information on the molecular mechanisms underlying such events.
Introduction
It is largely known that wound healing in animals is a fundamental process to re-establish the integrity and normal function of tissue layers after injury1. Despite epithelial surfaces exposed to the external environment (e.g., the skin, respiratory, and gastrointestinal tracts) form a protective barrier from physical and chemical insults, the formation of wounds can easily occur, especially after surgery or microbial infections2. As an example, colonization of lung tissue by the opportunistic bacterial pathogen Pseudomonas aeruginosa, especially in cystic fibrosis (CF) sufferers, leads to damage of the airways epithelium with consequent respiratory failure3,4. Wound healing is a complex host repair mechanism to restore the normal architecture of an injured tissue5. It is characterized by initial inflammation, followed by a regeneration period encompassing epithelialization, angiogenesis, and tissue remodeling with collagen production and cell differentiation6,7,8. To ensure epithelial integrity and to control microbial proliferation, all living organisms produce defense molecules, including antimicrobial peptides (AMPs)9,10. The wound healing process is very difficult to simulate in vitro due to the lack of cell debris and complex interactions among different cell types. However, the in vitro ability of a peptide to accelerate the closure of a pseudo-wound by stimulating migration of epithelial cells is indicative of its ability to heal a compromised epithelium. Indeed, cell migration is a rate-limiting event in wound healing, and studying factors that can affect cell migration will help to target therapies for improved wound healing.
Here, a highly reproducible experimental assay is provided based on special silicone culture inserts to evaluate cell migration in vitro. It is based on the creation of a 500 μm gap (pseudo-wound) on a confluent cell monolayer. The cells at the edge of the artificial "wounded" field will start migrating into the cell-free area, forming new cell-cell contacts. The culture insert represents a new tool for fast wound healing experiments. Two reservoirs separated by a 500 μm wall are provided, and they can be properly placed into a 3-cm dish plate or in the well of a 12-well plate. Filling each compartment of the insert with a cell suspension allows cells to grow in each designated area until confluence, while removal of the insert will engender a clean cell-free gap of approximately 500 μm (the same width as the separation wall). A proper cell culture medium supplemented with a test compound can then be added into the dish plate/well. Afterwards, the gap closure can be visualized at different time intervals under an inverted microscope, preferably one equipped with a video-camera for image acquisition. Finally, measurement of changes in the cell-covered area by the web-based automated image analysis program will allow the quantification of the speed of wound closure and cell migration. Overall, this method is a step forward with respect to the classical assay, where a scratch is manually made by incising confluent cell monolayers with a sterile needle or a pipette tip11. Indeed, the last procedure can destroy the plastic bottom of the dish plate/well and the surface coating, creating wrinkles. In addition, the "wounded" area does not have a well-defined width along the entire length of the gap, as this highly depends on the pressure applied by researchers to the needle/tip. Furthermore, the dislodged cells can form clumps of living and dead cells at the edges of the scratch; moreover, the spreading of living cells into the "wounded" area can interfere with the velocity of cell migration, generating non-reproducible results12.
In addition, thanks to a scratch image analysis platform, users can rapidly receive (within minutes) quantitative data on the migratory behavior of the selected cells without the necessity of acquiring additional software. This platform is capable of analyzing phase contrast microscopy images of low (~5X), medium (~10X), and high (~20X) magnification. After uploading a zip file of images (in *.jpg, *.jpeg, *.jp2, *.png, *.gif, *.tiff format) the analysis is automatically conducted to generate a summary file that shows the percentage of both cell-covered areas and scratch areas, as well as the speed of cell migration, at distinct time intervals.
In this work, by using a frog-skin AMP-derivative, i.e. Esc(1-21) and its diastereomer Esc(1-21)-1c13, and a bronchial cell line expressing the functional CF transmembrane conductance regulator (CFTR)14,15, an example of peptide-induced cell migration in comparison with untreated (control) samples is provided. Note that the airway epithelium and CFTR play a crucial role in maintaining lung function and wound repair16. Furthermore, by means of selective inhibitors (e.g., AG1478)17 of the epidermal growth factor receptor (EGFR), evidence that migration of bronchial cells induced by the aforementioned peptides involves activation of EGFR12,18 is reported.
In summary, the goal of this procedure is to show how the usage of such silicone culture inserts represents a fast and easily accessible assay to accurately determine migration of adherent cells (e.g., bronchial epithelial cells) and the molecular mechanisms controlling such events.
Protocol
1. Cell Preparation
- Seed 2.5x106 cells in 10 mL of Minimum Essential Medium (MEM) supplemented with 2 mM glutamine (MEMg), plus 10% fetal bovine serum (FBS), antibiotics (0.1 mg/mL of penicillin and streptomycin), and puromycin (0.5 µg/mL for selection and maintenance of the cell line) in a T75 flask. Incubate the flask at 37 °C and 5% CO2 for 2 days. Before starting the experiment, check the confluence of cells under an inverted microscope.
NOTE: The cells used for the experiment are immortalized human bronchial epithelial cells transduced with a lentiviral vector conferring resistance to puromycin. They stably express a functional CFTR14,15. - Once the cells' confluence has reached 90-100%, aspirate the medium from the flask and discard it into a waste bottle under a biological safety cabinet class II. Wash the cells with 6 mL of phosphate buffered saline without calcium and magnesium (CMF-PBS). Gently rock the flask manually and discard CMF-PBS.
NOTE: Be careful not to touch the cell monolayer with the pipette. - Add 10 mL of CMF-PBS and incubate the flask at 37 °C and 5% CO2 for 10 min.
- Aspirate CMF-PBS and discard it. Then add 2 mL of trypsin/EDTA to the flask.
- Gently rock the flask, allowing the solution to completely coat the cells, and incubate the flask at 37 °C and 5% CO2 for 10 min until the cells are visibly detached under a microscope.
NOTE: At the end of the incubation time, the cells should appear rounded and not attached to the plastic surface. If the cells are not well detached, manual agitation may be necessary. - Add 10 mL of MEMg plus 10% FBS to inactivate trypsin and collect the cells by washing the bottom of the flask. Transfer the volume into a conical 50 mL tube.
- Centrifuge the tube for 5 min at 80 x g.
- Aspirate the supernatant and re-suspend the cells in 6 mL of MEMg plus 10% FBS. Pipette repeatedly to break up any clumps.
- Take out 10 µL of cell suspension with a micropipette and inject the volume under the cover glass previously put over a Burker or Neubauer chamber.
- Count the cell number.
2. Cell Seeding in the Culture Inserts
- In each well of a 12-well plate, transfer the culture insert with sterile tweezers (Figure 1). Use tweezers to press along the inserts edges in order to fix them to the surface of the plate.
NOTE: Inserts have a sticky underside that allows adhesion.
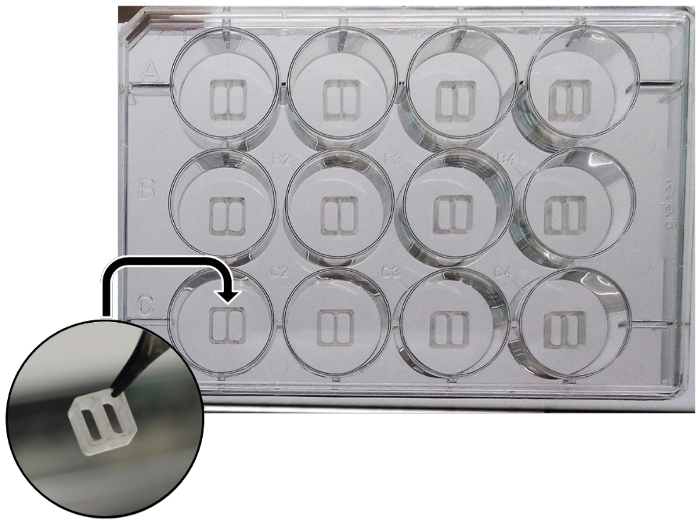
Figure 1: Schematic representation of the silicone culture inserts, properly put into wells of a 12-well plate. Please click here to view a larger version of this figure.
- Properly dilute the cell suspension in MEMg plus 10% FBS. Fill each compartment of the insert with 70 µL of cell suspension (about 3.5x104 cells/chamber).
NOTE: The density of cells applied in each compartment depends on the type of cells. It is recommended to use a cell density that leads to complete confluence within 24 h. - Under the microscope, check that cells are not leaking from the insert compartments and incubate the 12-well plate for 24 h at 37 °C and 5% CO2.
3. Pseudo-Wound Healing Assay
- After incubation, visualize the cells under the inverted microscope to verify that confluent cell monolayers have been formed.
- Re-suspend the test compounds (e.g., AMPs) in 1 mL of MEMg.
NOTE: Prepare fresh AMPs dilutions starting from the stock solution stored at -20 °C. - Gently remove the inserts by sterile tweezers. Be careful not to break the cell monolayers. Transfer the inserts onto absorbent paper.
NOTE: To re-use the same inserts, sterilize them in 70% ethanol for at least 3 h. It is recommended to throw them away afterwards. - To remove non-adherent cells, add 1 mL of MEMg per well using a micropipette. Close the plate and gently rock it.
NOTE: Do not add medium directly on top of cell monolayers to avoid their disruption. - Aspirate the medium and replace it with 1 mL of MEMg per well. Close the plate and visualize the cell-free gaps (created by the inserts) under the inverted microscope at 4X magnification, equipped with a video-camera. Acquire images at time zero (T0) and save them in a .jpg format.
- Remove the medium from the wells, wash them with 1 mL of PBS, and discard it.
- Add the test compounds (prepared at point 3.2) to the wells. For untreated control samples, add 1 mL of MEMg. Incubate the plate at 37 °C and 5% CO2.
- After 15, 20, and 24 h treatment, observe the cell migration under the microscope at 4X magnification and acquire images.
NOTE: During this step, try to capture images in the same areas as for T0. The choice of time intervals at which images are captured depends on the cell migration speed. - To study the effect of some selective inhibitors, i.e. AG1478, on cell migration, aspirate the medium from each insert compartment before removing inserts.
- Wash each compartment with fresh MEMg and fill it with 70 µL of MEMg supplemented with AG1478.
- After 30 min of incubation at 37 °C and 5% CO2, proceed as described from point 3.3.
NOTE: During the washing step and pretreatment of cell monolayers with the specific inhibitors, be careful not to remove the inserts.
4. Image Analysis
- Upon completion of the experiment, select images of the most representative samples of the various experimental groups, and create a zip file containing single images at T0, T15, T20, and T24 h.
NOTE: Single images are taken at the selected time intervals for all samples. Run triplicates for each experiment, which is repeated at least three times. At the end, for all experimental groups, a minimum of 3 images ("a", "b", "c", etc., deriving from each independent experiment) at each time point are analyzed. - Upload the zip file into the image analysis software which automatically provides by e-mail a spreadsheet summary file containing the experimental data of cell-covered and scratch areas (as percentage) at the selected time points.
NOTE: The recognition of the leading edge and the gap area is largely based on edge detection method (aimed at identifying points at which the image brightness sharply changes). - Save the data and collect them. Normalize all data with respect to the mean value at time zero. Calculate the average value of the normalized data of all replicates at each time point and the relative standard error (SEM). By using two-way analysis of variance (ANOVA), perform statistical analysis with a proper statistics software. Differences between peptide-treated groups and control groups at different time intervals are considered to be statistically significant for a p<0.05.
- Plot the obtained data into a graph as a histogram, which shows the percentage of cell-covered area of all sample groups versus the selected time intervals.
Results
This protocol was used to determine the wound healing effect of Esc(1-21) and Esc(1-21)-1c in terms of cell migration activity induced on bronchial epithelial cells expressing the functional CFTR. In this assay, culture inserts were placed in wells of a 12-well plate, and each compartment was seeded with 35,000 cells in MEMg supplemented with 10% FBS. The cells reached complete confluence within 24 h. Afterwards, a 500 μm gap was generated, and each well was filled with MEMg containi...
Discussion
Cell migration is an essential process in many physiological and pathological events including wound healing, embryonic development, and cancer metastasis. The basic procedure to study cell migration in vitro involves: (i) the creation of a cell monolayer, (ii) the production of a pseudo-wound in the confluent layer of cells, (iii) the capture of images at different time intervals until wound closure is reached, and (iv) the analysis of the image sequence in order to quantify the migration speed of the chosen ce...
Disclosures
The authors have nothing to disclose
Acknowledgements
This work was supported by funding from Sapienza University of Rome and the Italian Cystic Fibrosis Research Foundation (Project FFC#11/2014 adopted by FFC Delegations from Siena, Sondrio Valchiavenna, Cerea Il Sorriso di Jenny, and Pavia). Part of this work was also supported by FILAS Grant Prot. FILAS-RU-2014-1020.
We are grateful to Dr. Loretta Ferrera (U.O.C. Genetica Medica, Istituto Giannina Gaslini, Genova, Italy) for providing the bronchial epithelial cells.
Materials
| Name | Company | Catalog Number | Comments |
| Minimum essential medium (MEM) | Euroclone | ECB2071L | Warm in 37 °C water bath before use |
| Glutamine | Euroclone | ECB3000D | |
| Heat inactivated Fetal Bovine Serum (FBS) | Euroclone | ECS0180DH | |
| Penicillin and Streptomycin | Euroclone | ECB3001D | |
| Puromycin | Sigma-Aldrich | P8833 | |
| Trypsin/EDTA 1X in PBS | Euroclone | ECB3052D | Warm at room temperature before use |
| DPBS without calcium and magnesium (CMF-PBS) | Sigma-Aldrich | D8537 | |
| DPBS with calcium and magnesium (PBS) | Sigma-Aldrich | D8662 | |
| Ibidi Culture-Insert 2 well | Ibidi | 80209 | |
| Wimasis Image Analysis | Ibidi | 30002 | |
| PRISM software | GraphPad | version 6.0 |
References
- Enyedi, B., Niethammer, P. Mechanisms of epithelial wound detection. Trends Cell Biol. 25, 398-407 (2015).
- Kujath, P., Kujath, C. Complicated skin, skin structure and soft tissue infections - are we threatened by multi-resistant pathogens?. Eur J Med Res. 15, 544-553 (2010).
- Moreau-Marquis, S., Stanton, B. A., O'Toole, G. A. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther. 21, 595-599 (2008).
- Chiappini, E., Taccetti, G., de Martino, M. Bacterial lung infections in cystic fibrosis patients: an update. Pediatr Infect Dis J. 33, 653-654 (2014).
- Mangoni, M. L., McDermott, A. M., Zasloff, M. Antimicrobial peptides and wound healing: biological and therapeutic considerations. Exp Dermatol. 25, 167-173 (2016).
- Lau, K., Paus, R., Tiede, S., Day, P., Bayat, A. Exploring the role of stem cells in cutaneous wound healing. Exp Dermatol. 18, 921-933 (2009).
- Hu, M. S., et al. Tissue engineering and regenerative repair in wound healing. Ann Biomed Eng. 42, 1494-1507 (2014).
- Ramot, Y., et al. The role of PPARgamma-mediated signalling in skin biology and pathology: new targets and opportunities for clinical dermatology. Exp Dermatol. 24, 245-251 (2015).
- Lai, Y., Gallo, R. L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30, 131-141 (2009).
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature. 415, 389-395 (2002).
- Hulkower, K. I., Herber, R. L. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics. 3, 107-124 (2011).
- Di Grazia, A., et al. The frog skin-derived antimicrobial peptide esculentin-1a(1-21)NH2 promotes the migration of human HaCaT keratinocytes in an EGF receptor-dependent manner: a novel promoter of human skin wound healing?. PLoS One. 10, e0128663 (2015).
- Di Grazia, A., et al. D-Amino acids incorporation in the frog skin-derived peptide esculentin-1a(1-21)NH2 is beneficial for its multiple functions. Amino Acids. 47, 2505-2519 (2015).
- Cappiello, F., et al. Esculentin-1a-derived peptides promote clearance of Pseudomonas aeruginosa internalized in bronchial cells of cystic fibrosis patients and lung cell migration: biochemical properties and a plausible mode of action. Antimicrob Agents Chemother. 60, 7252-7262 (2016).
- Bebok, Z., et al. Failure of cAMP agonists to activate rescued deltaF508 CFTR in CFBE41o- airway epithelial monolayers. J Physiol. 569, 601-615 (2005).
- Trinh, N. T., et al. Improvement of defective cystic fibrosis airway epithelial wound repair after CFTR rescue. Eur Respir J. 40, 1390-1400 (2012).
- Gan, H. K., et al. The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J Biol Chem. 282, 2840-2850 (2007).
- Tokumaru, S., et al. Induction of keratinocyte migration via transactivation of the epidermal growth factor receptor by the antimicrobial peptide LL-37. J Immunol. 175, 4662-4668 (2005).
- Tjabringa, G. S., et al. The antimicrobial peptide LL-37 activates innate immunity at the airway epithelial surface by transactivation of the epidermal growth factor receptor. J Immunol. 171, 6690-6696 (2003).
- Di Grazia, A., Luca, V., Segev-Zarko, L. A., Shai, Y., Mangoni, M. L. Temporins A and B stimulate migration of HaCaT keratinocytes and kill intracellular Staphylococcus aureus. Antimicrob Agents Chemother. 58, 2520-2527 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved