A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Nanosensors to Detect Protease Activity In Vivo for Noninvasive Diagnostics
In This Article
Summary
Proteases are tightly regulated enzymes involved in fundamental biological processes, and dysregulated protease activity drives progression of complex diseases such as cancer. This method's goal is to create nanosensors that measure protease activity in vivo by producing a cleavage signal that is detectable from host urine and discriminates disease.
Abstract
Proteases are multi-functional enzymes that specialize in the hydrolysis of peptide-bonds and control broad biological processes including homeostasis and allostasis. Moreover, dysregulated protease activity drives pathogenesis and is a functional biomarker of diseases such as cancer; therefore, the ability to detect protease activity in vivo may provide clinically relevant information for biomedical diagnostics. The goal of this protocol is to create nanosensors that probe for protease activity in vivo by producing a quantifiable signal in urine. These protease nanosensors consist of two components: a nanoparticle and substrate. The nanoparticle functions to increase circulation half-life and substrate delivery to target disease sites. The substrate is a short peptide sequence (6-8 AA), which is designed to be specific to a target protease or group of proteases. The substrate is conjugated to the surface of the nanoparticle and is terminated by a reporter, such as a fluorescent marker, for detection. As dysregulated proteases cleave the peptide substrate, the reporter is filtered into urine for quantification as a biomarker of protease activity. Herein we describe construction of a nanosensor for matrix metalloproteinase 9 (MMP9), which is associated with tumor progression and metastasis, for detection of colorectal cancer in a mouse model.
Introduction
Proteases are multi-functional enzymes that specialize in the hydrolysis of peptide-bonds and have significant control over many biological processes, including homeostasis, allostasis, and disease1. An altered state of protease activity has been correlated to a variety of diseases, including cancer and cardiovascular disease, making proteases attractive candidates for development into clinical biomarkers2,3. Moreover, protease activity is functionally linked to distinct pathogeneses, patient outcomes, and prognosis of disease4. Broadly, biosensors have been developed to detect various biological phenomena and diseases, such as cancer, neurodegenerative disease, and electron transfer processes5,6,7,8,9. More specifically, substrate-based protease sensors have been developed to detect protease activity, and include fluorogenic probes for diagnostic imaging10 and isotopically labeled peptide substrates for in vitro detection by mass spectrometry11. In addition, activity-based probes have been developed, which contain substrate-like regions that bind or modify the target protease12. With this method, the target protease is irreversibly inhibited when the active site is modified, and analysis requires harvesting of tissue, which limits in vivo applications. However, it is important to sense protease activity in vivo, because regulation of protease activity is heavily dependent on the context of other biological activities such as the presence of endogenous inhibitors.
The goal of this work is to describe the formulation of activity-based nanosensors that detect protease activity in vivo by producing a measurable signal in urine. This platform is used as a noninvasive diagnostic to discriminate complex diseases such as cancer by using dysregulated protease activity as a functional biomarker. Our nanosensor platform consists of iron oxide nanoparticles (IONP) conjugated to protease substrates. These substrates are terminated by a fluorescent reporter which is released when proteases cleave the substrate. These IONPs circulate in vivo, localize to disease sites, and expose substrates to active disease-associated proteases. After cleavage, fluorescent reporters are released and, due to their small size, are filtered into urine, while uncleaved substrates on the IONP remain in the body. Therefore, an increase in protease activities in vivo will result in higher concentrations of reporter in urine (Figure 1). Since our platform is a urine test, no imaging platform is required and diagnostic signals are enriched in urine.
This platform can be engineered to detect a variety of diseases including cancer, fibrosis, and thrombosis13,14. Here we describe the design of nanosensors to detect elevations in Matrix metallopeptidase 9 (MMP9) activity as a biomarker of colorectal cancer. Colorectal cancer is the second leading cause of cancer death in the United States, with an estimated 136,800 new cases and 50,300 deaths in 2014 alone15. Colorectal tumor cells produce MMP9, which has been shown to drive malignant progression , matrix degradation, as well as metastasis16. Additionally, we identified a suitable peptide substrate (PLGVRGK) for MMP9 from the literature17. This platform may be used for early cancer detection and low-cost point-of-care diagnostics13,14,18,19,20,21.
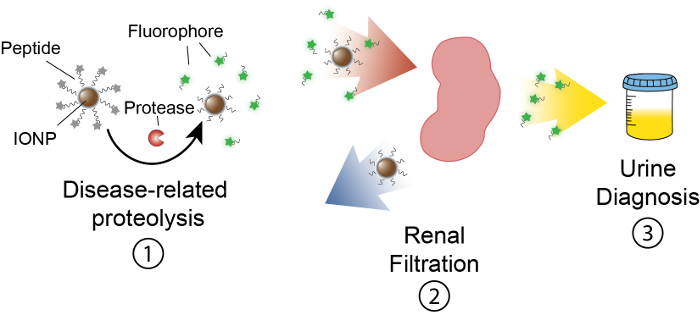
Figure 1: Schematic of Nanosensor Activity In vivo. Nanosensors circulate through the body and localize to sites of disease. Then, disease-related proteases cleave peptide substrates presented by IONPs. The size of cleaved fragments allows for renal clearance, causing them to localize in the urine. After the animal urinates, these peptide fragments can be analyzed by their reporter molecule. Please click here to view a larger version of this figure.
Protocol
Institutional approval from Institutional Animal Care and Use Committee (IACUC) at the researcher's institution is necessary to carry out the following animal experiments. Additionally, standard animal care facilities (e.g., housing chambers, sterile animal hoods, isofluorane chambers for anesthetization, and CO2 chambers for ethical endpoint euthanization) are necessary to properly carry out these experiments. Special training and assistance with these facilities can be provided by the Physiological Research Laboratory (PRL) at one's institution. All animal work was approved by IACUC at Georgia Tech (protocol: A14100).
1. Iron Oxide Nanoparticle (IONP) Synthesis
NOTE: Safety: The entire Iron Oxide Nanoparticle synthesis should be performed using personal protective equipment and underneath a chemical fume hood.
- In a 15 mL conical tube, chill 1 mL of 30% ammonium hydroxide in an ice bucket for 30 min.
- Wash a 250 mL Erlenmeyer flask with deionized water and add 10 mL double distilled water (ddH2O). Submerge the outside of the flask in an ice bath resting on a stir/heat plate. Use a plastic pipette to flow N2 gas into the ddH2O in the flask. Bubble for 15 minutes with N2 to deoxygenate.
- Weigh 224 µmol (4.5 grams) of dextran (Molecular Weight (MW) = 20 kDa) into a 50 mL conical tube. Add H2O such that the final volume of the mixture, including the dextran, is 20 mL. Vortex vigorously to dissolve the dextran.
- Weigh 290 µmol (78.5 grams) of iron(III) chloride hexahydrate, add to dextran solution and vortex to dissolve.
- Filter the resulting solution through a 0.2 µm filter.
- Transfer 1 mL chilled ammonium hydroxide into 9 mL of the deoxygenated water. Return to 4 °C.
- Weigh 367 µmol (73.0 grams) of iron(II) chloride tetrahydrate, dissolve in 1 mL of oxygen-free ddH2O, and filter through a 0.2 µm filter.
- Transfer the dextran-iron(III) solution into the 250 mL Erlenmeyer flask and deoxygenate with N2 for 15 minutes on ice. Mix with a magnetic stir bar spinning at 1600 rpm.
- To create a homogenous nitrogen atmosphere, cap the flask with a rubber septum. Puncture the septum with an 18 gauge needle to flow N2 into the flask. Insert a separate 18 gauge needle as a flow outlet.
- Add 467 µL of the iron(II) solution to the dextran-iron(III) solution with a 1 mL syringe, stirring with a magnetic stir bar at 1600 rpm. This ratio of iron(II) to iron(III) results in a balanced reaction that will produce magnetite, or Fe3O4.
- Add the chilled dilute ammonium hydroxide dropwise into the Iron(II)-Iron(III)-dextran solution to initiate the nucleation process21. Make sure each droplet mixes well before dropping in another. Finish the reaction after the addition of 1-2 mL of ammonium hydroxide.
- Stop the flow of N2 and remove the rubber septum. Remove the ice bath and replace with a bath of warm water, while continuing to stir the solution. Ensure that the temperature of the solution reaches 75 °C and incubate for 75 minutes.
- Remove the flask from the stir/hot plate and doubly filter the solution through 0.2 µm then 0.1 µm filters to remove coarse particles.
- Buffer exchange the particles into ddH2O, using 100 kDa molecular weight cut off (m.w.c.o.) concentrators to remove excess dextran from the solution, which should be very viscous. Replace filters if the flow through remains dark even after 2 - 3 spins, which indicates that the filter may have broken.
- To buffer exchange with spin filters, centrifuge at 4 °C at 4800 X g for 15 minutes, discard the flow-through, and add new buffer to the IONP solution. Repeat 3-5 times.
- Determine the concentration of the nanoparticles using a spectrophotometer/plate reader. Take absorbance measurements at 400 nm and use the molar absorptivity of IONP at this wavelength (ε = 2.07 x 106 cm-1 M-1) to determine the concentration of IO particles.
- Resuspend the IONPs to 10 mg/mL in ddH2O (usually total volume is ~3 mL) and make sure to use polypropylene tubes for this step for chemical compatibility. Add 1.6 volumes of 5 M NaOH, then add 0.65 volumes of epichlorohydrin. Rigorously mix on a plate shaker at room temperature for 12 hours to crosslink dextran.
- Use a 20 mL syringe with an 18-gauge needle to transfer the IONP solution into the 50 kDa m.w.c.o. dialysis membrane. Place the dialysis membrane into 4 L ddH2O and replace H2O a few times (every 2 - 3 hours). Incubate overnight.
- Measure the concentration and bring IONPs to 5 mg/mL (see step 1.15). Add ammonium hydroxide to reach 20% (v/v) and shake at room temperature for > 12 hours to aminate the surface of IONPs. Buffer exchange using 30 kDa m.w.c.o. filters (see 1.14).
- Adjust the volume down to 2.5 - 3.5 mL before final purification by Fast Protein Liquid Chromatography (FPLC; see Table of Materials for gel filtration column).
- Use dynamic light scattering (DLS) to determine the hydrodynamic radius of IONPs (expected range 10 - 100 nm, average size 40 - 50 nm). Do this by aliquoting 1 mL of 100X diluted IONP sample in a cuvette, place in the machine, and use the manufacturer's software to take the measurement.
NOTE: The total population of IONPs will be between 10 and 100 nm by DLS measurements, but the majority of the population is around the average diameter, which ranges from 40 - 50 nm. If one wants to limit the size range, one can further use size exclusion chromatography to isolate fractions with different diameters. IONPs should be stored at 4 °C.
2. Peptide Design, Conjugation to IONP, and In Vitro Validation
- Synthesize a peptide substrate (e.g., by a core facility or commercially) for a target protease with an N-terminal fluorescent reporter such as Fluorescein isothiocyanate (FITC) and a C terminal cysteine residue to allow thiol-mediated coupling.
NOTE: In the case of this study for MMP9, the peptide used was FITC-PLGVRGK-C, with kcat/Km ~ 2.0 x 105 M-1s-1. - Aliquot 0.5 mg of IONP (at 1 mg/mL), and buffer exchange into coupling buffer (50 mM Sodium Borate with 1 mM EDTA, pH = 8.5) using a 10 kDa m.w.c.o. spin filter (see step 1.14).
- Dissolve Succinimidyl iodoacetate (SIA) in dimethylformamide (DMF) to reach a concentration of ~30 mg/mL, and add SIA to IONP at a mole ratio of 500 SIA:IONP.
NOTE: SIA is a heterobifunctional cross linker that mediates coupling of amines on IONPs to the sulfhydryl groups on the peptide substrate. - Incubate for 1 - 2 hours at room temperature in the dark. If leaving overnight, incubate at 4 °C.
- Buffer exchange using a 10 kDa m.w.c.o. spin filter into coupling buffer to remove unreacted SIA (see step 1.14).
- Bring the final product solution to 1 mg/mL (0.5 mL solution). Mix the peptide of interest at a molar ratio of 90:1 (peptide:IONP) with 20 kDa thiol-terminated polyethylene glycol (PEG) at a molar ratio of 20:1 (PEG:IONP). Mix this peptide-PEG solution with IONP.
- Incubate overnight at room temperature on a plate shaker on highest speed, covering tubes in foil to prevent photobleaching of fluorescent molecules.
- Add L-cysteine at a molar ratio of 500:1 (C:IONP) to neutralize any unreacted SIA molecules. Incubate for 1 hour at 25 °C on a shaker at highest speed at room temperature.
- Purify via FPLC (see 1.1.19). Use a spectrophotometer to measure the absorbance of the sample solution and calculate the Peptide:IONP ratio according to the following. After purification, store product at 1 mg/mL at 4 °C in PBS.
- Use a spectrophotometer to measure the absorbance of the sample solution at 400 nm (Asample,400) and the wavelength of maximum absorbance for the fluorophore used, which is 488 nm for FITC (Asample,488). Measure the absorbance of a stock amine IONP solution at the same two wavelengths (400 nm and 488 nm), which are respectively represented as ANP,400 and ANP,488.
- Use equations 1 and 2 to calculate the normalized A400 and A488 values.
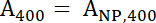 Equation 1
Equation 1
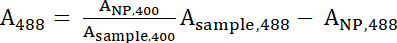 Equation 2
Equation 2 - A400 and A488 represent the normalized values of the sample. Use these values to calculate the ratio of Peptide:IONP with equation 3.
 Equation 3
Equation 3 - Where
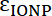 and
and 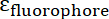 are the molar absorptivities of the IONP and the fluorophore, respectively. For these particles
are the molar absorptivities of the IONP and the fluorophore, respectively. For these particles 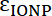 = 2.06 x 106 M-1cm-1 and for FITC
= 2.06 x 106 M-1cm-1 and for FITC 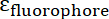 = 72,000 M-1cm-1, so the value of
= 72,000 M-1cm-1, so the value of  should be 28.75. Typical Peptide:IONP ratios should be in the range of 20:1 to 50:1.
should be 28.75. Typical Peptide:IONP ratios should be in the range of 20:1 to 50:1.
- Validate functionality of the probes by performing an in vitro cleavage assay.
- With PBS (1% BSA), make an 18 µL nanoparticle solution with a 200 nM concentration of peptide. Mix with 2 µL of protease of interest (MMP9; 0.1 - 1 mg/mL).
- Incubate in a plate reader at 37 °C for 1 hour, taking fluorescence measurements (using the appropriate excitation and emission wavelengths, which are 485 nm and 528 nm for FITC, respectively) every 1 - 2 minutes to monitor cleavage.
3. Administration of Nanosensors and Urine Detection of Cancer
NOTE: For more details on the example model, see our previous report13.
- Create a metabolic cage for urine collection by securing a cylindrical sleeve to the top of a 96 well plate. During urine collection, place a mouse inside the sleeve and cover with a Petri dish cover to prevent escape of the animal.
- Prepare an injection solution (200 µL maximum volume) containing nanosensors at a concentration of 3000 mg/kg (~50 µmol) by peptide in sterile saline.
- In female, 8-week old, nude mice bearing xenograft LS174T colorectal tumors at a burden of ~100-300 mm3, which should occur on approximately day 10, administer nanosensor solution via tail vein injection. Immediately after injection, place mice into metabolic cages and note the time of injection for each mouse.
- At 60 - 90 minutes after injection, remove cylindrical sleeve from 96-well plate. Restrain mouse and apply slight pressure on the bladder to induce mice to void any remaining urine onto the plate. Collect all urine (200 - 500 µL).
- Analyze urine samples by immunoprecipitation to purify FITC from the urine and increase sensitivity. Use magnetic beads coupled with anti-FITC antibodies.
- First, wash 25 mg of the magnetic beads 3 times with coating buffer, and bring the final volume to 225 µL.
NOTE: It is important to make the coating buffer (0.1 M sodium borate, pH 9.5), blocking buffer (PBS, 0.5% BSA, 0.05% Tween-20, pH 7.4), and washing buffer (PBS, 0.1% BSA, 0.05% Tween-20, pH 7.4) fresh each time, and approximately 20 mL of each is necessary. - Add 200 µL of anti-FITC (5 mg/mL) and 200 µL of ammonium sulfate (3 M). Incubate for 16-24 hours at 37 C on a rotator.
- Add the solution to a magnetic separator, remove the supernatant, replace with 625
 Λ blocking buffer, and incubate at 37 C overnight. Then wash 3 times with wash buffer and store in 1.25 mL wash buffer.
Λ blocking buffer, and incubate at 37 C overnight. Then wash 3 times with wash buffer and store in 1.25 mL wash buffer. - Incubate 2 µL of urine with 5 µL of magnetic beads (20 mg/mL) and bring to a total volume to 50 µL with PBS (0.01% Tween 20). Incubate for 60 min.
- Wash twice with 50 µL PBS (0.01% Tween 20) using a magnetic separator to collect magnetic beads after each wash.
- Elute twice with 32.5 µL of 5% glacial Acetic Acid. Neutralize pooled elution (70 µL) with 35 µL of 2 M Tris to achieve a final pH of ~7.
- Read on a plate reader (see Table of Materials) at appropriate excitation and emission wavelengths to quantify urine fluorescence. Calculate the concentration of fluorescent reporter against a ladder of known concentrations of free fluorophore.
- First, wash 25 mg of the magnetic beads 3 times with coating buffer, and bring the final volume to 225 µL.
Results
The majority of the population of the IONPs is around the average diameter, which ranges from 40 - 50 nm. After pegylation, this size range has a circulation half-life of approximately 6 hours13 in vivo (see Figure 2a). If one wants to select for a particular size range, one can use size exclusion chromatography to isolate IONP fractions with different diameters. The nanoparticles by TEM will appear as individual spherica...
Discussion
This method describes the development of activity-based nanosensors consisting of protease substrates conjugated to a nanoparticle core. The event of proteolytic cleavage is dubbed the "pharmacokinetic switch", because cleaved peptide products are smaller than the renal size filtration limit of 5 nm23 and filter into urine to produce a noninvasive signal. Therefore, it is important to use nanoparticles or carriers with a hydrodynamic radius that is larger than 5 nm, as anything smalle...
Disclosures
Dr. Kwong is co-founder and serves as consultant to Glympse Bio, which is developing products related to the research described in this paper. This study could affect his personal financial status. The terms of this arrangement have been reviewed and approved by Georgia Tech in accordance with its conflict of interest policies
Acknowledgements
This work was funded by an NIH Director's New Innovator Award (Award No. DP2HD091793). Q.D.M. is supported by the NSF Graduate Research Fellowships Program (Grant No. DGE-1650044). B.A.H is supported by the National Institutes of Health GT BioMAT Training Grant under Award Number 5T32EB006343 as well as the Georgia Tech President's Fellowship. G.A.K. holds a Career Award at the Scientific Interface from the Burroughs Welcome Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| Name | Company | Catalog Number | Comments |
| 0.2 µm syringe filters | VWR | 4652 | |
| 18G needle | VWR | 89134-024 | |
| 15 mL conicals | VWR | 89039-670 | |
| 250 mL Erlenmeyer flask | VWR | 89000-362 | |
| Stir bar | VWR | 58949-006 | |
| Hot Plate/Magnetic Stirrer | VWR | 97042-634 | |
| Glacial acetic acid | VWR | 97064-482 | |
| Albumin from Bovine Serum (BSA) | Thermo Fisher | A13100 | |
| Iron (III) chloride hexahydrate | Sigma | 236489 | |
| Iron (II) chloride tetrahydrate | Sigma | 44939 | |
| Epichlorohydrin | Sigma | 45340-500ML-F | |
| DMF | Sigma | D4551 | |
| Ammonium Hydroxide | Sigma | 320145-500ML | |
| Sodium Hydroxide pellets | Sigma | 221465-500G | |
| EDTA | Sigma | E9884 | |
| Sodium Borate | Sigma | B9876 | |
| L-Cysteine | Sigma | 168149-100G | |
| Tris-HCl | Sigma | T5941 | |
| Tris base | Sigma | T6066 | |
| PBS tablets | Sigma | P4417 | |
| Dextran | Pharmacosmos | 5510 0020 9006 | |
| Amicon 15 mL 10k filters, 24 pk | Millipore | UFC901024 | |
| Amicon 15 mL 30k filters, 24 pk | Millipore | UFC903024 | |
| Amicon 15 mL 100k filters, 24 pk | Millipore | UFC910024 | |
| Zetasizer Nano ZS | Malvern Panalytical | NanoZS | |
| Slide-A-Lyzer Dialysis Cassette | LifeTech | 66130 | |
| Dynabeads MyOne Tosylactivated | LifeTech | 65501 | |
| SIA | Life Tech | 22349 | |
| PEG 20k | Laysan Bio | MPEG-SH-20K-1g | |
| Fluorescein Antibody [2A3] | GeneTex | GTX10257 | |
| Hiload 16/600 superdex 200 | GE Healthcare | 45-002-490 | |
| Plate Reader | Fisher | BTCYT5M | |
| BD Insulin Syringes | Fisher | NC0872854 |
References
- Lopez-Otin, C., Bond, J. S. Proteases: Multifunctional enzymes in life and disease. Journal of Biological Chemistry. 283 (45), 30433-30437 (2008).
- Hua, Y., Nair, S. Proteases in cardiometabolic diseases: Pathophysiology, molecular mechanisms and clinical applications. Biochimica et Biophysica Acta. 1852 (2), 195-208 (2015).
- Koblinski, J. E., Ahram, M., Sloane, B. F. Unraveling the role of proteases in cancer. Clinica Chimica Acta. 291 (2), 113-135 (2000).
- Lilja, H., Vickers, A., Scardino, P. Measurements of proteases or protease system components in blood to enhance prediction of disease risk or outcome in possible cancer. Journal of Clinical Oncology. 25 (4), 347-348 (2007).
- Jin, H., et al. Flexible surface acoustic wave resonators built on disposable plastic film for electronics and lab-on-a-chip applications. Scientific Reports. 3, 2140 (2013).
- Ma, W., Liu, H. -. T., Long, Y. -. T. Monitoring dopamine quinone-induced dopaminergic neurotoxicity using dopamine functionalized quantum dots. ACS Applied Materials & Interfaces. 7 (26), 14352-14358 (2015).
- Zhang, W. -. H., Ma, W., Long, Y. -. T. Redox-mediated indirect fluorescence immunoassay for the detection of disease biomarkers using dopamine-functionalized quantum dots. Analytical Chemistry. 88 (10), 5131-5136 (2016).
- Ma, W., et al. Investigating electron-transfer processes using a biomimetic hybrid bilayer membrane system. Nature Protocols. 8 (3), 439-450 (2013).
- Holt, B. A., et al. Fc microparticles can modulate the physical extent and magnitude of complement activity. Biomaterials science . 5, 463-474 (2017).
- Edgington, L. E., Verdoes, M., Bogyo, M. Functional imaging of proteases: Recent advances in the design and application of substrate-based and activity-based probes. Current Opinion in Chemical Biology. 15 (6), 798-805 (2011).
- Kleifeld, O., et al. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nature Protocols. 6 (10), 1578-1611 (2011).
- Sanman, L. E., Bogyo, M. Activity-based profiling of proteases. Annual Review of Biochemistry. 83 (1), 249-273 (2014).
- Kwong, G. A., et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nature Biotechnology. 31 (1), 63-70 (2013).
- Dudani, J. S., Buss, C. G., Akana, R. T. K., Kwong, G. A., Bhatia, S. N. Sustained-release synthetic biomarkers for monitoring thrombosis and inflammation using point-of-care compatible readouts. Advanced Functional Materials. 26 (17), 2919-2928 (2016).
- Meester, R. G. S., et al. Public health impact of achieving 80% colorectal cancer screening rates in the United States by 2018. Cancer. 121 (13), 2281-2285 (2015).
- Mehner, C., et al. Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget. 5 (9), 2736-2749 (2014).
- Bremer, C., Tung, C. H., Weissleder, R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nature Medicine. 7 (6), 743-748 (2001).
- Warren, A. D., et al. Disease detection by ultrasensitive quantification of microdosed synthetic urinary biomarkers. Journal of the American Chemical Society. 136 (39), 13709-13714 (2014).
- Warren, A. D., Kwong, G. A., Wood, D. K., Lin, K. Y., Bhatia, S. N. Point-of-care diagnostics for noncommunicable diseases using synthetic urinary biomarkers and paper microfluidics. Proceedings of the National Academy of Sciences of the United States of America. 111 (10), 3671-3676 (2014).
- Kwong, G. A., et al. Mathematical framework for activity-based cancer biomarkers. Proceedings of the National Academy of Sciences of the United States of America. 112 (41), 12627-12632 (2015).
- Dudani, J. S., Jain, P. K., Kwong, G. A., Stevens, K. R., Bhatia, S. N. Photoactivated spatiotemporally-responsive nanosensors of in vivo protease activity. ACS Nano. 9 (12), 11708-11717 (2015).
- Palmacci, S., Josephson, L. Synthesis of polysaccharide covered superparamagnetic oxide colloids. US patent. , (1991).
- Choi, H. S., et al. Renal clearance of nanoparticles. Nature Biotechnology. 25 (10), 1165-1170 (2007).
- Harris, T. J., von Maltzahn, G., Derfus, A. M., Ruoslahti, E., Bhatia, S. N. Proteolytic actuation of nanoparticle self-assembly. Angewandte Chemie (International edition in English). 45 (19), 3161-3165 (2006).
- Kwon, E. J., Dudani, J. S., Bhatia, S. N. Ultrasensitive tumour-penetrating nanosensors of protease activity. Nature Biomedical Engineering. 1, (2017).
- Villanueva, J., Nazarian, A., Lawlor, K., Tempst, P. Monitoring Peptidase activities in complex proteomes by MALDI-TOF mass spectrometry. Nature Protocols. 4 (8), 1167-1183 (2009).
- Villanueva, J., et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. The Journal of Clinical Investigation. 116 (1), 271-284 (2006).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved