A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Development of Targeting Induced Local Lesions IN Genomes (TILLING) Populations in Small Grain Crops by Ethyl Methanesulfonate Mutagenesis
In This Article
Summary
Described is a protocol for developing a Targeting Induced Local Lesions IN Genomes (TILLING) population in small grain crops with use of ethyl methanesulfonate (EMS) as a mutagen. Also provided is a protocol for mutation detection using the Cel-1 assay.
Abstract
Targeting Induced Local Lesions IN Genomes (TILLING) is a powerful reverse genetics tool that includes chemical mutagenesis and detection of sequence variation in target genes. TILLING is a highly valuable functional genomics tool for gene validation, especially in small grains in which transformation-based approaches hold serious limitations. Developing a robust mutagenized population is key to determining the efficiency of a TILLING-based gene validation study. A TILLING population with a low overall mutation frequency indicates that an impractically large population must be screened to find desired mutations, whereas a high mutagen concentration leads to high mortality in the population, leading to an insufficient number of mutagenized individuals. Once an effective population is developed, there are multiple ways to detect mutations in a gene of interest, and the choice of platform depends upon the experimental scale and availability of resources. The Cel-1 assay and agarose gel-based approach for mutant identification is convenient, reproducible, and a less resource-intensive platform. It is advantageous in that it is simple, requiring no computational knowledge, and it is especially suitable for validation of a small number of genes with basic lab equipment. In the present article, described are the methods for development of a good TILLING population, including preparation of the dosage curve, mutagenesis and maintenance of the mutant population, and screening of the mutant population using the PCR-based Cel-1 assay.
Introduction
Point mutations in genomes can serve many useful purposes for researchers. Depending on their nature and location, these mutations can be used to assign functions to genes or even distinct domains of proteins of interest. On the other hand, as a source of novel genetic variation, useful mutations can be selected for desired traits using phenotyping screens and further used in crop improvement. TILLING is a powerful reverse genetics tool that includes chemical mutagenesis and detection of sequence variation in the target gene. First developed in Arabidopsis1 and Drosophilia melanogaster2, TILLING populations have been developed and utilized in many small grain crops such as hexaploid bread wheat (Triticum aestivum)3, barley (Hordeum vulgare)4, tetraploid durum wheat (T. dicoccoides durum)5, diploid wheat (T. monococcum)6 and the "D" genome progenitor of wheat Aegilops tauschii7. These resources have been used to validate the roles of genes in regulating abiotic and biotic stress tolerance8, regulating flowering time9, and developing nutritionally superior crop varieties5.
TILLING, along with the use of alkylating mutagenic agents such as ethyl methanesulfonate (EMS), sodium azide, N-methyl-N-nitrosourea (MNU), and methyl methanesulfonate (MMS), has advantages over other reverse genetics tools for several reasons. First, mutagenesis can be conducted on practically any species or variety of plant10 and is independent of the transformation bottleneck, which is particularly challenging in the case of small grains11. Second, in addition to generating knockout mutations that can be obtained by other gene validation approaches, a range of missense and splicing mutations can be induced, which can discern functions of individual domains of the proteins of interest12. Moreover, TILLING generates an immortal collection of mutations throughout the genome; thus, a single population can be used for functional validation of multiple genes. In contrast, other reverse genetics tools generate resources specific to only the gene under study13. Useful mutations identified through TILLING can be deployed for breeding purposes and are not subject to regulation, unlike gene editing, whose non-transgenic classification is still uncertain in many countries. This becomes especially relevant to small grains that are internationally traded14.
TILLING is a simple and efficient gene validation strategy and requires mutagenized populations to be developed for investigating genes of interest. Developing an effective mutagenized population is key to determining the efficiency of a TILLING-based gene validation study. A TILLING population with a low overall mutation frequency indicates that an impractically large population must be screened for desired mutations, whereas a high mutagen concentration leads to high mortality in the population and an insufficient number of mutagenized individuals. Once a good population is developed, there are multiple ways to detect mutations in the genes of interest, and the choice of platform depends on the experimental scale and availability of resources. Whole genome sequencing and exome sequencing has been used to characterize all mutations in TILLING populations in plants with small genomes15,16. Exome sequencing of two TILLING populations has been performed in bread and durum wheat and is available to the public for identifying desirable mutations and ordering mutant lines of interest17. It is a great public resource in terms of availability of desirable mutations; however, in gene validation studies, the wild-type line should possess the candidate gene of interest. Unfortunately, it is still cost-prohibitive to sequence the exome of the entire TILLING population for reverse genetics-based validation of a few candidate genes in another background. Amplicon sequencing and Cel-1-based assays have been used in detecting mutations in targeted populations in wheat, and Cel-1 assays are simpler, requiring no computational knowledge, and are especially suitable for validation of a small number of genes with basic lab equipment6,18.
In the present article, described are methods for the development of a good TILLING population, including preparation of the dosage curve, mutagenesis and maintenance of the mutant population, and screening of the mutant population using the PCR-based Cel-1 assay. This protocol has already been implemented successfully in developing and utilizing mutagenized populations of Triticum aestivum, Triticum monoccocum6, barley, Aegilops tauchii7, and several others. Included are explicit details of these methods along with useful tips that will help researchers develop TILLING populations, using EMS as a mutagen in any small grain plant of choice.
Protocol
1. Preparation of dosage curve for effective mutagenesis
- Soak 100 seeds with the genotype of interest in six 250 mL glass flasks (100 in each flask) containing 50 mL of distilled water. Shake at 100 rpm for 8 h at room temperature (RT) for imbibition by the seeds.
- In a fume hood, prepare 50 mL of 0.4%, 0.6%, 0.8%, 1.0%, and 1.2% (w/v) ethyl methanesulfonate (EMS) solution by dissolving 0.167, 0.249, 0.331, 0.415, and 0.498 mL of EMS in distilled water, respectively.
NOTE: EMS is liquid at RT with a density of 1.206 g/mL.
CAUTION: Use appropriate personal protective equipment (PPE) while handling EMS. - Decant the water out of five flasks and add 50 mL of EMS solution in each flask containing imbibed seed so that there are six different treatments with 0.0%, 0.4%, 0.6%, 0.8%, 1.0%, and 1.2% EMS solution. Shake flasks for 16 h at 75 rpm and RT.
- Decant the EMS solution and collect the treated seeds separately for each treatment using cheese cloth. Inactivate the used EMS solution by adding one volume of EMS-inactivating solution (0.1 M NaOH, 20% w/v Na2S2O3) for 24 h. Also treat the contaminated flasks and pipette tips with the EMS-inactivating solution for 24 h.
- Wash the EMS-treated seeds under running tap water for 2 h. Transplant each seed individually into root trainers containing potting soil.
- Grow plants at 20–25 °C under a 16 h light period.
- Record data on plant survival after 15 days of transplantation. Use the following equation to calculate the survival rate for each treatment:
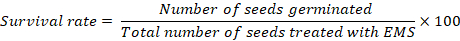
NOTE: If the germination rate is lower than 100% in controls, accurate survival rates in all the treatments should be calculated after subtracting the number of seeds that failed to germinate in the controls. A survival rate of 40%–60% is desirable for effective mutagenesis. It may be required to perform a second round of dosage optimization with a modified concentration according to the survival of the treated seeds until achieving the desirable lethality rate of 40%–60%.
2. Mutagenesis and maintenance of mutant population
- Soak the final batch of at least 3,000 seeds dividing equally (600 seeds each) in five 1,000 mL flasks containing 300 mL distilled water. Shake for 8 h at 100 rpm under RT for imbibition.
NOTE: The final size of the desirable population will depend on the mutation frequency and ploidy level of the genotype, but it is advisable to use at least 3,000 seeds for hexaploids, 4,000 seeds for tetraploids, and more than 7,000 seeds for diploids. - In a fume hood, prepare 1,500 mL of the optimized EMS concentration solution in distilled water.
- Decant the water out of the flasks and add 300 ml of the optimal concentration of EMS solution in each flask containing 600 imbibed seeds. Shake the flasks for 16 h at 75 rpm and RT.
- Decant the EMS and collect the treated seeds in cheese cloth. Inactivate the EMS solution and treatment containers with EMS-inactivating solution as done in step 1.4.
- Wash the EMS-treated seeds under running tap water for 2 h. Transplant each EMS-treated M1 seed individually into root trainers.
- Grow M1 plants (derived from M0 seeds) at 20–25 °C under a 16 h light period.
NOTE: It may be required to vernalize the seedlings at the two leaf stage for 6 weeks at 4 °C, if the genotype of interest has a winter-type growth habit. - Allow the M1 plants to self-pollinate, and harvest the M2 seeds separately for each fertile M1 plant.
NOTE: To avoid chances of potential outcrossing, cover the spikes of M1 plants with pollination bags before anthesis. - Plant a single M2 seed from each M1 plant to avoid genetic redundancy.
- Collect tissue from M2 plants at the two-leaf stage in 1.1 mL of racked 96 well microtubes. Collect around 80 mm of leaf tissue from each plant and record the ID of each sample in a tissue collection plan.
- Freeze-dry the leaf tissue using a lyophilizer and store at -80 °C.
- Maintain the M2 plants at 20–25 °C under a 16 h light period.
- Record data on mutant phenotypes of the M2 plants at regular intervals. The expected phenotypes are albino, chlorina, grassy shoot, variegated, partially fertile, sterile, etc.
- Allow the M2 plants to self-fertilize and mature. Separately harvest and save the M3 seeds of the M2 plants (Figure 1).
NOTE: M seeds are used for validating the phenotype in reverse genetics studies. Therefore, M should be carefully catalogued and stored under cool and dry conditions. Alternatively, if the seed must be increased in field, head rows can be planted for each M plant, and M seeds from each row can be harvested and saved separately as the TILLING population resource.
3. Cel-1 assay for genetic characterization of mutants
- Extract DNA from the leaf tissue of M2 using a plant DNA extraction kit with a DNA purification system (see Table of Materials) following the manufacturer’s recommendations.
- Quantify DNA using a spectrophotometer and normalize the DNA concentrations to 25 ng/µL with nuclease-free water in 96 well blocks.
NOTE: It is important to check the quality of the DNA by running it on a gel, as low-quality DNA (smeared DNA) may result in false negatives in pooled samples. - Create 4x DNA pools by combining DNA from four 96 well blocks into one plate, while maintaining the row and column identity of each sample. Add 50 µL of DNA from each individual sample into the pool plate so that each pool plate well contains a total 200 µL of DNA from four different 96 well blocks.
- Catalogue the identity of pooled DNA in the format of Pool Plate-Row-Column (e.g., Pool 1 A1 = Box1A1 + Box2A1 + Box3A1 + Box4A1).
- Design genome-specific primers [A1] for the gene of interest using the Genome Specific Primers (GSP) page <https://probes.pw.usda.gov/GSP> with default settings for polyploid species. For diploids, use Primer 3 <http://primer3.ut.ee/> for designing primers using default settings. Design multiple primers, if needed, to cover the entire coding region of the gene of interest.
NOTE: The latest IWGSC assembly can be used to obtain sequences for the gene of interest in wheat using the URGI BLAST tool <https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST>. The optimal amplicon length is in the range of 800–1,500 bp. Table 1 shows examples of primers for waxy genes in hexaploid wheat. - Run a PCR for gene-specific primers on pooled DNA as follows:
- Add 5 µL of PCR buffer, 2 µL (each) of 4 µM forward and reverse primers, 0.1 µL of DNA polymerase (see Table of Materials), 5 µL of pooled DNA template, then increase the volume to 25 µL using nuclease-free water.
- Use a touch down profile to run the PCR reaction on a thermal cycler as follows: 95 °C for 1 min, seven cycles of 95 °C for 1 min, 67 °C to 60 °C for 1 min with temperature decreases of 1 °C per cycle and 72 °C for 2 min, followed by 30 cycles of 95 °C for 1 min, 60 °C for 1 min, 72 °C for 2 min, and final extension at 72 °C for 7 min.
NOTE: Above PCR profile works on wheat DNA template for most of the primers designed using primer 3 default settings. In case of non-specific amplification, PCR profile should be made stringent before moving to the next steps.
- Generate heteroduplexes between mismatched DNA by incubating PCR products in thermal cycler using profile as follows: 95 °C for 2 min, five cycles of 95 °C for 1 s, 95 °C to 85 °C for 1 min with temperature decreases of 2 °C per cycle, and 60 cycles of 85 °C to 25 °C with decreases of 1 °C per cycle.
- Add 2.5 µL of homemade Cel-1 endonuclease to the heteroduplexed PCR products and incubate for 45 min at 45 °C. Terminate the Cel-1 reaction by adding 2.5 µL of 0.5 M EDTA (pH 8.0).
NOTE: Cel-1 endonuclease can be extracted from fresh celery stalks using the protocol performed by Till et al.19 It is very important to test the activity and optimum amount of Cel-1 endonuclease, which can be tested using previously characterized mutants or a commercially available mutation detection kit. - Run the Cel-1 treated products on a 3.0% agarose gel at 100 V for 2.5 h. The wells containing smaller and unique cleaved band(s), in addition to full-length uncleaved bands, will contain the mutant DNA sample.
- Denconvolute mutant pools.
- Follow steps 3.6, 3.7, 3.8, and 3.9 for individual M2 DNA samples constituting the mutant pools identified in step 3.9.
- To determine the zygosity of mutants, run two PCR (as described in step 3.6) for individual M2 DNA, in which the first reaction contains 2.5 µL of M2 DNA and 2.5 µL of wild-type DNA and the second reaction contains only 5 µL of M2 DNA. If the mutation is heterozygous, an additional cleaved band will be present in both reactions. On the other hand, additional cleaved bands will be found only in the first reaction if the mutant is homozygous.
- To identify the nature of the mutation, sequence the PCR products of the confirmed mutants using a Sanger sequencing platform following the manufacturer’s instructions.
4. Calculation of mutation frequency
NOTE: Mutation frequency of a TILLING population refers to the average physical distance in which one mutation occurs in the individuals of that population. For example, a mutation frequency of 1/35 kb in a TILLING population means that an average individual of that population possesses 1 mutation per every 35 kb in the genome.
- To determine the mutation frequency of a TILLING population, calculate the total number of bases screened.
- To calculate the total number of bases screened, multiply the PCR product size by the total number of individuals screened.
- Divide the total number of bases screened by the number of unique mutations observed using the following equation, which will yield the physical region possessing 1 mutation in the given TILLING population:
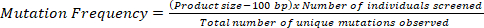
NOTE: To account for the limitation in resolution of 50 bp at both the ends based on an agarose gel-based platform, subtract 100 bp from the product size in the calculation.
Results
Figure 2 shows the dosage curve of hexaploid bread wheat cultivar Jagger, diploid wheat Triticum monococcum6, and a genome donor of wheat Aegilops tauschii7. The EMS doses for desired 50% survival rates were about 0.25%, 0.6% and 0.7% for T. monococcum, Ae. tauschii, and T. aestivum, respectively. The higher EMS tolerance of hexaploid wheat is due to its genome buffer...
Discussion
TILLING is a highly valuable reverse genetics tool for gene validation, especially for small grains where transformation-based approaches have serious bottlenecks11. Developing a mutagenized population with a high mutation frequency is one of the critical steps in conducting functional genomics studies. The most important step in developing a robust TILLING population is to determine the optimal concentration of EMS. The 40%-60% survival rate in the M1 has been found to be a good indica...
Disclosures
Authors declare no competing financial interests.
Acknowledgements
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project 1016879 and Maryland Agricultural Experiment Station via MAES Grant No. 2956952.
Materials
| Name | Company | Catalog Number | Comments |
| 96 well 1.1 ml microtubes in microracks | National Scientific | TN0946-08R | For collecting leaf tissues |
| Agarose I biotechnology grade | VWR | 0710-500G | |
| Biosprint 96 DNA Plant Kit | Qiagen | 941558 | Kit for DNA extraction |
| Cel-1 endonuclease | Extracted as described by Till et al 2006 | Single strand specific endonuclease | |
| Centrifuge 5430 R | Eppendorf | ||
| Ethyl methanesulfonate | Sigma Aldrich | M-0880-25G | EMS, Chemical mutagen |
| Freeze Dry/Shell freeze system | Labconco | For lyophilization of leaf tissue | |
| Kingfisher Flex purification system | Thermo fisher scientific | 5400610 | High throughput DNA extraction robot |
| My Taq DNA Polymerase | Bioline | BIO-21107 | |
| Nuclease free water | Sigma aldrich | W4502-1L | |
| NuGenius gel imaging system | Syngene | ||
| Orbit Environ-shaker | Lab-line | ||
| SPECTROstar Nano | BMG LABTECH | Nano drop for DNA quantification | |
| T100 Thermal cycler | BIO-RAD | 1861096 |
References
- McCallum, C. M., Comai, L., Greene, E. A., Henikoff, S. Targeted screening for induced mutations. Nature Biotechnology. 18 (4), 455-457 (2000).
- Bentley, A., MacLennan, B., Calvo, J., Dearolf, C. R. Targeted Recovery of Mutations in Drosophila. Genetics. 156 (3), 1169-1173 (2000).
- Tsai, H., et al. Discovery of Rare Mutations in Populations: TILLING by Sequencing. Plant Physiology. 156 (3), 1257-1268 (2011).
- Caldwell, D. G., et al. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). The Plant Journal. 40 (1), 143-150 (2004).
- Hazard, B., et al. Induced Mutations in the Starch Branching Enzyme II ( SBEII ) Genes Increase Amylose and Resistant Starch Content in Durum Wheat. Crop Science. 52 (4), 1754-1766 (2012).
- Rawat, N., et al. A diploid wheat TILLING resource for wheat functional genomics. BMC Plant Biology. 12, 205 (2012).
- Rawat, N., et al. TILL-D: An Aegilops tauschii TILLING Resource for Wheat Improvement. Frontiers in Plant Science. 9, (2018).
- Rawat, N., et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nature Genetics. 48 (12), 1576-1580 (2016).
- Kippes, N., Chen, A., Zhang, X., Lukaszewski, A. J., Dubcovsky, J. Development and characterization of a spring hexaploid wheat line with no functional VRN2 genes. Theoretical and Applied Genetics. 129 (7), 1417-1428 (2016).
- Greene, E. A., et al. Spectrum of Chemically Induced Mutations From a Large-Scale Reverse-Genetic Screen in Arabidopsis. Genetics. 164 (2), 731-740 (2003).
- Harwood, W. A. Advances and remaining challenges in the transformation of barley and wheat. Journal of Experimental Botany. 63 (5), 1791-1798 (2012).
- Henikoff, S., Comai, L. Single-Nucleotide Mutations for Plant Functional Genomics. Annual Review of Plant Biology. 54 (1), 375-401 (2003).
- Uauy, C., et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biology. 9 (1), 115 (2009).
- Uauy, C., Wulff, B. B. H., Dubcovsky, J. Combining Traditional Mutagenesis with New High-Throughput Sequencing and Genome Editing to Reveal Hidden Variation in Polyploid Wheat. Annual Review of Genetics. 51 (1), 435-454 (2017).
- Li, G., et al. The Sequences of 1504 Mutants in the Model Rice Variety Kitaake Facilitate Rapid Functional Genomic Studies. The Plant Cell. 29 (6), 1218-1231 (2017).
- Jiao, Y., et al. A Sorghum Mutant Resource as an Efficient Platform for Gene Discovery in Grasses. The Plant Cell. 28 (7), 1551-1562 (2016).
- Krasileva, K. V., et al. Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences. , 201619268 (2017).
- Dong, C., Dalton-Morgan, J., Vincent, K., Sharp, P. A Modified TILLING Method for Wheat Breeding. The Plant Genome. 2 (1), 39-47 (2009).
- Till, B. J., Zerr, T., Comai, L., Henikoff, S. A protocol for TILLING and Ecotilling in plants and animals. Nature Protocols. 1 (5), 2465-2477 (2006).
- Wu, J. -. L., et al. Chemical- and Irradiation-induced Mutants of Indica Rice IR64 for Forward and Reverse Genetics. Plant Molecular Biology. 59 (1), 85-97 (2005).
- Feldman, M., Levy, A. A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics. 192 (3), 763-774 (2012).
- Comai, L. The advantages and disadvantages of being polyploid. Nature Reviews Genetics. 6 (11), 836-846 (2005).
- Guo, H., et al. Development of a High-Efficient Mutation Resource with Phenotypic Variation in Hexaploid Winter Wheat and Identification of Novel Alleles in the TaAGP.L-B1 Gene. Frontiers in Plant Science. 8, (2017).
- Rakszegi, M., et al. Diversity of agronomic and morphological traits in a mutant population of bread wheat studied in the Healthgrain program. Euphytica. 174 (3), 409-421 (2010).
- Tsai, H., Ngo, K., Lieberman, M., Missirian, V., Comai, L. Tilling by Sequencing. Plant Functional Genomics: Methods and Protocols. , 359-380 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved