A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
A Proinflammatory, Degenerative Organ Culture Model to Simulate Early-Stage Intervertebral Disc Disease.
* These authors contributed equally
In This Article
Summary
This protocol presents a novel experimental model of proinflammatory, degenerative bovine organ culture to simulate early-stage intervertebral disc degeneration.
Abstract
Symptomatic intervertebral disc (IVD) degeneration (IDD) is a major socioeconomic burden and is characterized by inflammation and tissue degradation. Due to the lack of causative therapies, there is an urgent need for innovative experimental organ culture models to study the mechanisms involved in the progression of the disease, find therapeutic targets, and reduce the need for animal models. We here present a novel, three-dimensional organ culture model protocol mimicking the proinflammatory and catabolic microenvironment, which is present during IDD.
Initially, bovine caudal IVDs were dissected, cleaned, and cultured in the tissue culture medium. Dynamic physiologic or pathologic loading was applied in a custom-made bioreactor for 2 hours per day. IVDs were assigned to a control group (high glucose medium, physiological loading, phosphate-buffered saline injection) and a pathological group (low glucose medium, pathological loading, tumor necrosis factor-alpha injection) for four days. Gene expression analysis from collected nucleus pulposus cells of the IVDs and enzyme-linked immunosorbent assay of the conditioned organ culture media was performed.
Our data revealed a higher expression of inflammatory markers and reduced disc heights after loading in the pathological group compared to the control group. This protocol is reliable to simulate IVD inflammation and degeneration and can be further expanded to broaden its application scope.
Introduction
Low back pain (LBP) can affect individuals of all ages and is a leading cause for disability worldwide1,2,3. The total cost associated with LBP exceeds $100 billion per year4,5. Symptomatic intervertebral disc (IVD) degeneration (IDD), a condition characterized by inflammation and tissue degradation, is a major cause of LBP6,7. Specifically, IDD is characterized by a gradually evolving breakdown of the IVD's extracellular matrix (ECM), induced and triggered by multiple factors that lead to an accelerated pathology, neurological disorders, and eventually disability. Furthermore, IDD is associated with the release of proinflammatory cytokines, altered spine biomechanics, angiogenesis, and nerve ingrowth, which increases pain sensation, altogether causing chronic LBP (active discopathy)6,8. To date, treatment options include discectomy and subsequent fusion of the adjacent vertebrae, implantation of an IVD prosthesis, or non-surgical approaches, such as non-steroidal anti-inflammatory drugs, opioids, and muscle relaxants for patients with IDD9. Both current standard therapeutic options, surgical and non-surgical, are only partly effective and fail to address the underlying biological problem9,10. Early-stage degenerative disc disease is characterized by an initial inflammatory tissue response, especially an increase in tumor necrosis factor-alpha (TNF-alpha) expression11. These early disc changes primarily occur at the cellular level without disrupting the disc architecture and could previously be mimicked by nutritional deficiency under pro-inflammatory conditions12. Therefore, precise simulation of the in vivo situation to investigate these degeneration mechanisms and find suitable therapeutic targets is crucial. Additionally, to these simulations of molecular properties, the mechanical loading environment of the discs plays a key role in pathological and physiological changes of IVD. Consequently, combining these approaches would bring us one step forward to mimic the complex microenvironment of IVDs in vivo. There are currently no studies considering the aspect of dynamic loading along with the pro-inflammatory and nutritional setting to the best of our knowledge.
Although large animal models allow the investigation of potential relevant in vivo interactions, they are costly and work intensive. Moreover, as the use of animal models in research has long been a matter of controversy, the reduction of the number of animals needed to answer important research questions is of great interest. Finally, there is currently no ideal animal model to mimic IDD in IVD research13,14. Therefore, it is necessary to establish a cost-effective and reliable replacement, such as an organ culture model to simulate IDD and associated inflammatory and degenerative processes. Recently, the application of the present protocol on the establishment of a proinflammatory and degenerative organ culture model to simulate early-stage intervertebral disc disease allowed us to investigate the effect of anti-inflammatory drugs in the IDD organ culture15.
Here, we describe how to obtain bovine intervertebral discs and induce the state of early-stage IDD via a catabolic and proinflammatory microenvironment caused by direct intradiscal injection of tumor necrosis factor-alpha (TNF-α) and degenerative loading in a bioreactor under low nutritive medium conditions. Figure 1 illustrates the experimental model and shows the bioreactor used to simulate degenerative and physiological loading conditions.
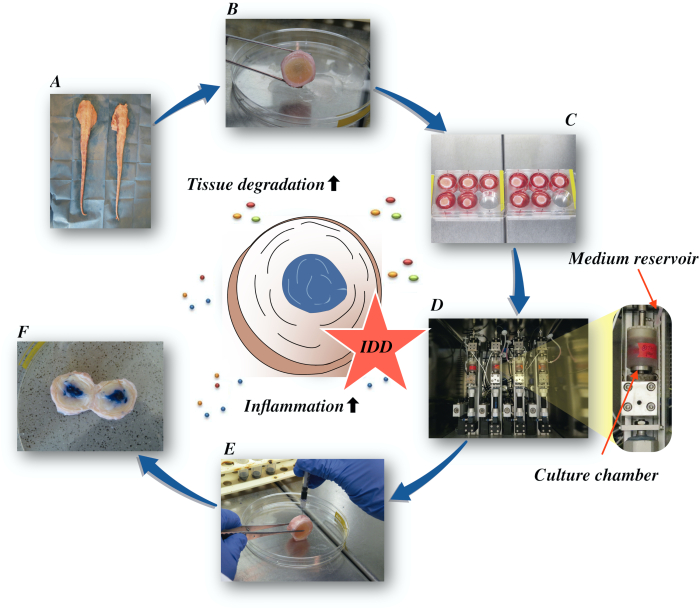
Figure 1: Illustration of the experimental setup. A: bovine tail; B: dissected bovine intervertebral discs; C: transfer of the disc to a well-plate with culture medium; D: loading the simulation in a bioreactor; E: intradiscal injection technique; F: IVD after injection of PBS/trypan blue dye to reveal distribution. IDD: intervertebral disc degeneration. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Protocol
Experiments were performed using bovine tails obtained from local abattoirs. The biological materials used in the current study are taken from the food chain and require no ethical approval in Swiss and European law.
1. Dissection of the bovine intervertebral disc
- Rinse the whole tail thoroughly with tap water to remove dirt and hair on the surface.
NOTE: With intact, distal ends, a maximum of 9 IVDs (coccygeal 1-9) per tail can be used for the experiments depending on the desired size of the IVDs. Considering the desired diameter between 15-20 mm, we used 12 bovine tails with 5 IVDs per tail for the experiments. - Immerse the whole tail in a box containing 1% betadine solution for 10 min. Briefly dry the tail with sterile gauze and place it on a sterile drape.
NOTE: During dissection of the disc, humidify the tails with Ringer's solution wetted gauze to prevent dehydration. Store the tails (or left-over segments) wrapped in wet gauze until the whole dissection procedure is completed. - Use a scalpel (No. 20) to remove the soft tissue as completely as possible from the caudal spine to facilitate the identification of the IVDs. Remove the spinous and transverse processes of the vertebrae with bone removal pliers.
NOTE: Select IVDs with the desired diameter. IVDs with a diameter range of 15-20 mm were used in the current study. - Cut transversely with bone pliers through the middle of each vertebral body to obtain individual motion segments. Put motion segments in a Petri dish with gauze wetted with Ringer's solution.
- Locate the IVD and vertebra by palpation and by moving the motion segments gently. Make two parallel cuts with the band saw in the growth plate of the IVDs, one on each side of the IVD. Identify the location of the growth plate by touching and finding the convex site of the bony endplate part (hard) adjacent to the disc (soft) with a safety distance of approximately 0.5-1 mm from the IVD towards the vertebra. Ensure that the blade of the band saw is cooled with Ringer's solution while cutting the vertebrae.
- Transfer IVDs in a clean Petri dish with clean gauze wetted with Ringer's solution.
NOTE: The gauze should be moistened and not too wet to prevent swelling of the IVDs, - Use the scalpel blade to scrape off the vertebral body (red/pink bone), growth plate (white cartilage), leave the endplate intact (yellow-pink). Make the two surfaces flat and parallel for the loading procedure. Transfer scraped IVDs to a fresh Petri dish with gauze wetted with Ringer's solution.
NOTE: Wear a chainmail glove to protect the hand while holding the IVD and scraping. - Measure the disc height and diameter with a caliper. Clean the blood clots in the vertebrae bone with Ringer's solution using a jet lavage system.
- Transfer the IVDs to 50 mL plastic tubes, one IVD per tube. Add 25 mL of Phosphate-Buffered Saline (PBS) + 10% Penicillin/Streptomycin (P/S) per IVD and leave it shaking for 15 min on an orbital shaker at room temperature.
- Aspirate the supernatant and add 10 mL of PBS + 1% P/S per IVD for 2 min to rinse the IVDs.
2. IVD culture and loading
- Transfer discs to IVD chambers and add IVD culture medium (Dulbecco's Modified Eagle Medium (DMEM, 4.5 g/L high glucose DMEM for the physiological group and 2 g/L low glucose DMEM for the pathological group) + 1% P/S + 2% fetal calf serum + 1% ITS (contains 5 µg/mL insulin, 6 µg/mL transferrin, and 5 ng/mL selenious acid) + 50 µg/mL ascorbate-2-phosphate + 1% non-essential amino acid + 50 µg/mL antimicrobial agent for primary cells) and place in an incubator at 37 °C, 85% humidity and 5% CO2.
- Culture the discs for 4 days within a bioreactor system according to experimental groups16. In the pathologic group, maintain degenerative loading conditions at 0.32-0.5 MPa, 5 Hz for 2 h/day. In the physiological control group, use a loading protocol of 0.02-0.2 MPa, 0.2 Hz for 2 h/day.
NOTE: Position the IVDs in chambers containing 5 mL of IVD medium during the loading procedures. The volume depends on the size of the bioreactor's loading chambers. Between the loading procedures, place the IVDs in six-well plates with 7 mL of IVD culture medium for free-swelling recovery. - For analyzing the changes in disc height during the experimental period, measure the disc height with a caliper after IVD dissection (baseline) and then daily after the free swelling period and after dynamic loading for the experimental duration.
3. Intradiscal tumor necrosis factor-alpha (TNF-α) injection
- Directly after the first dynamic loading cycle on day 1, place the IVDs in a Petri dish in a vertical position and stabilize the IVDs with a tweezer.
- Inject recombinant TNF-α (100 ng in 70 µL of PBS per IVD) with a 30-gauge insulin needle into the nucleus pulposus tissue of the pathological group17. Inject slowly at a speed of approximately 70 µL in 1 min.
- After injection, pull the syringe halfway back within the IVD and pull the syringe plunger to create a vacuum that prevents the injected solution from leaking back, before removing the needle and syringe completely from the IVD.
NOTE: Perform a pilot experiment by injecting PBS containing trypan blue dye to evaluate the distribution of the injected solution after loading and overnight culture.
4. Gene expression
- Harvest the IVDs on day 4. Collect the nucleus pulposus (NP) tissue (gelly part in the middle of IVD) with a biopsy punch. Collect the outer annulus fibrosus (AF) with a scalpel blade (No.20).
NOTE: For the baseline reference at day 0, collect tissues immediately after dissection for RNA extraction. - Use the amount of NP or AF tissue needed for gene expression analysis, depending on the experimental design.
NOTE: For the present experiments, approximately 150 mg tissue was used. The ratio of RNA isolation solution to tissue mass should be at least 2 mL per 100-150 mg tissue for efficient extraction. - Digest the NP or AF tissue with the digestion solution (0.2% pronase in DMEM, filter sterilized) and incubate for 1 h at 37 °C with magnetic stirring18.
- Flash-freeze the tissue samples using liquid nitrogen and pulverize to a fine powder. Divide the pulverized tissue powder equally into two 2 mL tubes each containing 1 mL of guanidine thiocyanate and phenol in a monophase solution (RNA isolation solution).
- Perform the homogenization in 2 mL tubes containing the RNA isolation solution and the pulverized tissue powder. Homogenize the tissue powder 5x with an 8 mm stainless steel ball and a tissue-lyzer at 30 Hz for 3 min. Centrifuge at 12,000 x g, 4 °C for 10 min and transfer the supernatant to a fresh tube. The supernatants can be stored at -80 °C for at least one month.
- Add 0.1 mL of 1-bromo-3-chloropropane (BCP) per 1 mL of RNA isolation solution and shake vigorously for 15 s. Store the resulting mixture at room temperature on an orbital shaker for 15 min and centrifuge at 12,000 x g for 15 min at 4 °C.
NOTE: The RNA remains exclusively in the upper aqueous phase. - Transfer the aqueous phase into a fresh tube and precipitate RNA with 0.25 mL of isopropanol and 0.25 mL of high salt precipitation solution per 1 mL of RNA isolation solution used for initial homogenization. Store the samples at room temperature for 15 min on an orbital shaker and centrifuge at 12,000 x g for 8 min at 4 °C.
NOTE: Alternatively, use the column-based RNA extraction method which generally leads to higher RNA purity but lower RNA yield. - Remove the supernatant and wash the RNA pellet with 1 mL of 75% ethanol per 1 mL of RNA isolation solution used for initial homogenization. Centrifuge at 7,500 x g for 5 min at 4 °C.
- Remove the ethanol wash and briefly air-dry RNA pellet for 3-5 min. Dissolve the RNA in 20 µL of diethylpyrocarbonate (DEPC) treated water by passing the solution a few times through a pipette tip and incubating for 10-15 min at 55-60 °C.
- Measure the absorbance at 230 nm, 260 nm, and 280 nm (A230, A260 and A280 respectively). A260 of 1.0 corresponds to 40 µg/mL RNA. An A260/A280 ratio of 1.6-1.9 is expected, whereas contamination results in a A260/A280 ratio of <1.6.
- Prepare a reverse transcriptase (RT) reaction mix for a 20 µL reaction volume. The mix contains RT enzyme mix, ribonuclease inhibitor, helper protein, primers, dNTPs, MgCl2, RNase free water, and 0.4 μg of RNA sample.
- Briefly centrifuge the RT tubes to mix all the components at the bottom of the tube.
- Place the samples in the thermocycler instrument. Select the appropriate program for the RT. Run the RT for 10 min at 25 °C, followed by the reverse transcription step for 120 min at 42 °C and inactivation of the reverse transcriptase for 5 min at 85 °C, cooling it down to 4 °C at the end.
- Dilute the resulting cDNA with Tris(hydroxymethyl)aminomethane (Tris)-ethylenediaminetetraacetic acid (EDTA) (TE) buffer (10 mM Tris with 1 mM EDTA) to a final concentration of 0.4 µg RNA used for RT per 100 µL cDNA solution. Store the cDNA samples at -20 °C.
- Perform real-time polymerase chain reaction (PCR) using a 10 µL reaction volume. The reaction volume contains the master mix (containing DNA polymerase, uracil-DNA glycosylase, dNTPs with dUTP, passive reference and optimized buffer components), forward primer 45 µM, reverse primer 45 µM, probe 12.5 µM (containing a reporter dye linked to the 5' end of the probe, a minor groove binder at the 3' end of the probe, and a nonfluorescent quencher at the 3' end of the probe), 2 µL cDNA and DEPC-treated water.
- Run an endogenous control (RPLP0) for relative quantification with the 2-ΔΔCT method19.
- Add samples in duplicates and run a no template control by adding TE-buffer instead of cDNA. Run PCR at standard conditions (2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C).
- Perform relative quantification of mRNA targets following the comparative CT method. The amount of mRNA normalized to the baseline sample is calculated as 2-ΔΔCT, whereas ΔΔCT is the difference between the ΔCT (CT target- CT endogenous control) of sample and ΔCT (CT target and CT endogenous control) of the baseline sample.
5. Quantification of protein content in the IVD medium
- Collect the medium conditioned by the IVD samples to measure the protein content in the medium. Perform enzyme-linked immunosorbent assay (ELISA) according to the protocol of the target protein.
- Bovine interleukine-8 (IL-8) is quantified by anti-bovine IL8 ELISA kit according to the manufacturer's instruction.
Access restricted. Please log in or start a trial to view this content.
Results
Degenerative loading in low glucose medium combined with TNF-α injection caused a significant increase of the gene expression of proinflammatory markers interleukin 6 (IL-6) and interleukin 8 (IL-8) compared to the physiological control group in NP cells after 4 days of culture (Figure 2). In contrast, we did not observe significant changes for the proinflammatory genes interleukin 1β (IL-1β) and TNF-α in NP cells (data not shown). Furthermore, degenerative culture condit...
Access restricted. Please log in or start a trial to view this content.
Discussion
We here provided a detailed protocol to simulate degenerative and inflammatory IVDD. This protocol can be applied for detailed examinations of inflammatory pathways leading to the destructive effects on the disc. Moreover, the protocol can help to determine promising therapeutic targets involved in the progression of the disease.
We recently showed that human recombinant TNF-α could induce inflammation in both bovine and human NP cells21, which is in accordance wit...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by AO Foundation and AOSpine International. Babak Saravi received fellowship support from the German Spine Foundation and the German Osteoarthritis Foundation. Gernot Lang was supported by the Berta-Ottenstein-Programme for Advanced Clinician Scientists, Faculty of Medicine, University of Freiburg, Germany.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 1-Bromo-3-chloropropane(BCP) | Sigma-Aldrich, St. Louis, USA | B9673 | |
| Ascorbate-2-phosphate | Sigma-Aldrich, St. Louis, USA | A8960 | |
| Band saw | Exakt Apparatebau, Norderstedt, Germany | model 30/833 | |
| Betadine | Munndipharma, Frankfurt, Germany | ||
| Bovine IL-8 Do.it-Yourself ELISA | Kingfisher Biotech, St. Paul, USA | DIY1028B-003 | |
| Corning ITS Premix | Corning Inc., New York, USA | 354350 | |
| DMEM high glucose | Gibco by life technologies, Carlsbad, USA | 10741574 | |
| DMEM low glucose | Gibco by life technologies, Carlsbad, USA | 11564446 | |
| Ethanol for molecular biology | Sigma-Aldrich, St. Louis, USA | 09-0851 | |
| Fetal Bovine Serum (FBS) | Gibco by life technologies, Carlsbad, USA | A4766801 | |
| Non-essential amino acid solution | Gibco by life technologies, Carlsbad, USA | 11140050 | |
| Penicillin/Streptomycin(P/S) | gibco by life technologies, Carlsbad, USA | 11548876 | |
| Phosphate Buffer Solution, tablet | Sigma-Aldrich, St. Louis, USA | P4417 | |
| Pronase | Sigma-Aldrich, St. Louis, USA | 10165921001 | |
| Primocin | InvivoGen, Sandiego, USA | ant-pm-05 | |
| Pulsavac Jet Lavage System | Zimmer, IN,USA | ||
| TissueLyser II | Quiagen, Venlo, Netherlands | 85300 | |
| Streptavidinn-HRP | Kingfisher Biotech, St. Paul, USA | AR0068-001 | |
| Superscript VILO | Invitrogen by life Technologies, Carlsbad, USA | 10704274 | |
| cDNA Synthesis Kit | Applied Biosystems by life technologies | 10400745 | |
| TaqMan Universal Master Mix | Applied Biosystems by life technologies | ||
| TNF-alpha, recombinant human protein | R&D systems, Minnesota, USA | 210-TA-005 | |
| TRI Reagent | Molecular Research Center, Cincinnati, USA | TR 118 | |
| Tris-EDTA buffer solution | sigma-Aldrich, St. Louis, USA | 93283 | |
| Gene bIL-6 | Applied Biosystems by life technologies | Custom made probes | Primer fw (5′–3′) TTC CAA AAA TGG AGG AAA AGG A Primer rev (5′–3′) TCC AGA AGA CCA GCA GTG GTT Probe (5′FAM/3′TAMRA) CTT CCA ATC TGG GTT CAA TCA GGC GATT |
| Gene bIL8 | Applied Biosystems by life technologies | Bt03211906_m1 | |
| Gene bTNF-alpha | Applied Biosystems by life technologies | Custom made probes | Primer fw (5′–3′) CCT CTT CTC AAG CCT CAA GTA ACA A Primer rev (5′–3′) GAG CTG CCC CGG AGA GTT Probe (5′FAM/3′TAMRA) ATG TCG GCT ACA ACG TGG GCT ACC G |
| GENE bIL1beta | Applied Biosystems by life technologies | Custom made probes | Primer fw (5′–3′) TTA CTA CAG TGA CGA GAA TGA GCT GTT Primer rev (5′–3′) GGT CCA GGT GTT GGA TGC A Probe (5′FAM/3′TAMRA) CTC TTC ATC TGT TTA GGG TCA TCA GCC TCA A |
| RPLP0 | Applied Biosystems by life technologies | Bt03218086_m1 |
References
- Vos, T., et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet. 390 (10100), 1211-1259 (2017).
- Hoy, D., et al. Measuring the global burden of low back pain. Best Practice & Research Clinical Rheumatology. 24 (2), 155-165 (2010).
- Thiese, M. S., et al. Prevalence of low back pain by anatomic location and intensity in an occupational population. BMC Musculoskeletal Disorders. 15 (1), 283(2014).
- Katz, J. N. Lumbar Disc Disorders and Low-Back Pain: Socioeconomic Factors and Consequences. The Journal of Bone and Joint Surgery (American). 88, suppl_2 21(2006).
- Vlaeyen, J. W. S., et al. Low back pain. Nature Reviews Disease Primers. 4 (1), 52(2018).
- Khan, A. N., et al. Inflammatory biomarkers of low back pain and disc degeneration: a review: Biomarkers of disc degeneration and back pain. Annals of the New York Academy of Sciences. 1410 (1), 68-84 (2017).
- Kim, H. S., Wu, P. H., Jang, I. T. Lumbar Degenerative Disease Part 1: Anatomy and Pathophysiology of Intervertebral Discogenic Pain and Radiofrequency Ablation of Basivertebral and Sinuvertebral Nerve Treatment for Chronic Discogenic Back Pain: A Prospective Case Series and Review of Literature. International Journal of Molecular Sciences. 21 (4), 1483(2020).
- Adams, M. A., Roughley, P. J. What is Intervertebral Disc Degeneration, and What Causes It. Spine. 31 (18), 2151-2161 (2006).
- Wu, P. H., Kim, H. S., Jang, I. T. Intervertebral Disc Diseases Part 2: A Review of the Current Diagnostic and Treatment Strategies for Intervertebral Disc Disease. International Journal of Molecular Sciences. 21 (6), 2135(2020).
- Lurie, J. D., et al. Surgical Versus Nonoperative Treatment for Lumbar Disc Herniation: Eight-Year Results for the Spine Patient Outcomes Research Trial. Spine. 39 (1), 3-16 (2014).
- Risbud, M. V., Shapiro, I. M. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nature Reviews Rheumatology. 10 (1), 44-56 (2014).
- Ponnappan, R. K., et al. An organ culture system to model early degenerative changes of the intervertebral disc. Arthritis Research & Therapy. 13 (5), 171(2011).
- O'Connell, G. D., Vresilovic, E. J., Elliott, D. M. Comparison of Animals Used in Disc Research to Human Lumbar Disc Geometry. Spine. 32 (3), 328-333 (2007).
- Stannard, J. T., et al. Development of a whole organ culture model for intervertebral disc disease. Journal of Orthopaedic Translation. 5, 1-8 (2016).
- Li, Z., et al. Preclinical ex-vivo Testing of Anti-inflammatory Drugs in a Bovine Intervertebral Degenerative Disc Model. Frontiers in Bioengineering and Biotechnology. 8, 583(2020).
- Li, Z., et al. Development of an ex vivo cavity model to study repair strategies in loaded intervertebral discs. European Spine Journal. 25 (9), 2898-2908 (2016).
- Kazezian, Z., Li, Z., Alini, M., Grad, S., Pandit, A. Injectable hyaluronic acid down-regulates interferon signaling molecules, IGFBP3 and IFIT3 in the bovine intervertebral disc. Acta Biomaterialia. 52, 118-129 (2017).
- Caprez, S., Menzel, U., Li, Z., Grad, S., Alini, M., Peroglio, M. Isolation of high-quality RNA from intervertebral disc tissue via pronase predigestion and tissue pulverization. JOR Spine. 1 (2), 1017(2018).
- Lopa, S., Ceriani, C., Cecchinato, R., Zagra, L., Moretti, M., Colombini, A. Stability of housekeeping genes in human intervertebral disc, endplate and articular cartilage cells in multiple conditions for reliable transcriptional analysis. European Cells & Materials. 31, 395-406 (2016).
- Lang, G., et al. An intervertebral disc whole organ culture system to investigate proinflammatory and degenerative disc disease condition. Journal of Tissue Engineering and Regenerative Medicine. 12 (4), 2051-2061 (2018).
- Du, J., et al. Proinflammatory intervertebral disc cell and organ culture models induced by tumor necrosis factor alpha. JOR Spine. 3, 1104(2020).
- Purmessur, D., Walter, B. A., Roughley, P. J., Laudier, D. M., Hecht, A. C., Iatridis, J. A role for TNFα in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochemical and Biophysical Research Communications. 433 (1), 151-156 (2013).
- Walter, B. A., Likhitpanichkul, M., Illien-Junger, S., Roughley, P. J., Hecht, A. C., Iatridis, J. C. TNFα Transport Induced by Dynamic Loading Alters Biomechanics of Intact Intervertebral Discs. PLOS One. 10 (3), 0118358(2015).
- Gullbrand, S. E., et al. A large animal model that recapitulates the spectrum of human intervertebral disc degeneration. Osteoarthritis and Cartilage. 25 (1), 146-156 (2017).
- Willems, N., et al. Safety of intradiscal injection and biocompatibility of polyester amide microspheres in a canine model predisposed to intervertebral disc degeneration: intradiscal application of pea microspheres. Journal of Biomedical Materials Research Part B: Applied Biomaterials. 105 (4), 707-714 (2017).
- Michalek, A. J., Buckley, M. R., Bonassar, L. J., Cohen, I., Iatridis, J. C. The effects of needle puncture injury on microscale shear strain in the intervertebral disc annulus fibrosus. The Spine Journal. 10 (12), 1098-1105 (2010).
- Illien-Jünger, S., et al. The combined effects of limited nutrition and high-frequency loading on intervertebral discs with endplates. Spine. 35 (19), 1744-1752 (2010).
- Gantenbein, B., et al. Organ culture bioreactors--platforms to study human intervertebral disc degeneration and regenerative therapy. Current Stem Cell Research & Therapy. 10 (4), 339-352 (2015).
- Boubriak, O. A., Watson, N., Sivan, S. S., Stubbens, N., Urban, J. P. G. Factors regulating viable cell density in the intervertebral disc: blood supply in relation to disc height. Journal of Anatomy. 222 (3), 341-348 (2013).
- Maroudas, A., Stockwell, R. A., Nachemson, A., Urban, J. Factors involved in the nutrition of the human lumbar intervertebral disc: cellularity and diffusion of glucose in vitro. Journal of Anatomy. 120, Pt 1 113-130 (1975).
- Beckstein, J. C., Sen, S., Schaer, T. P., Vresilovic, E. J., Elliott, D. M. Comparison of Animal Discs Used in Disc Research to Human Lumbar Disc: Axial Compression Mechanics and Glycosaminoglycan Content. Spine. 33 (6), 166-173 (2008).
- Walter, B. A., Illien-Jünger, S., Nasser, P. R., Hecht, A. C., Iatridis, J. C. Development and validation of a bioreactor system for dynamic loading and mechanical characterization of whole human intervertebral discs in organ culture. Journal of Biomechanics. 47 (9), 2095-2101 (2014).
- Rajan, N. E., et al. Toll-Like Receptor 4 (TLR4) Expression and Stimulation in a Model of Intervertebral Disc Inflammation and Degeneration. Spine. 38 (16), 1343-1351 (2013).
- vanden Akker, G. G., Rorije, A. J., Davidson, E. N. B., vander Kraan, P. M. Phenotypic marker genes distinguish inner and outer annulus fibrosus from nucleus pulposus tissue in the bovine intervertebral disc. Osteoarthritis and Cartilage. 25, 402(2017).
- Du, J., et al. Functional cell phenotype induction with TGF-β1 and collagen-polyurethane scaffold for annulus fibrosus rupture repair. European Cells & Materials. 39, 1-17 (2020).
- Risbud, M. V., et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. Journal of Orthopaedic Research: Official Publication of the Orthopaedic Research Society. 33 (3), 283-293 (2015).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved