A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Visualization and Quantification of TGFβ/BMP/SMAD Signaling under Different Fluid Shear Stress Conditions using Proximity-Ligation-Assay
In This Article
Summary
Here, we establish a protocol to simultaneously visualize and analyze multiple SMAD complexes using proximity ligation assay (PLA) in endothelial cells exposed to pathological and physiological fluid shear stress conditions.
Abstract
Transforming Growth Factor β (TGFβ)/Bone Morphogenetic Protein (BMP) signaling is tightly regulated and balanced during the development and homeostasis of the vasculature system Therefore, deregulation in this signaling pathway results in severe vascular pathologies, such as pulmonary artery hypertension, hereditary hemorrhagic telangiectasia, and atherosclerosis. Endothelial cells (ECs), as the innermost layer of blood vessels, are constantly exposed to fluid shear stress (SS). Abnormal patterns of fluid SS have been shown to enhance TGFβ/BMP signaling, which, together with other stimuli, induce atherogenesis. In relation to this, atheroprone, low laminar SS was found to enhance TGFβ/BMP signaling while atheroprotective, high laminar SS, diminishes this signaling. To efficiently analyze the activation of these pathways, we designed a workflow to investigate the formation of transcription factor complexes under low laminar SS and high laminar SS conditions using a commercially available pneumatic pump system and proximity ligation assay (PLA).
Active TGFβ/BMP-signaling requires the formation of trimeric SMAD complexes consisting of two regulatory SMADs (R-SMAD); SMAD2/3 and SMAD1/5/8 for TGFβ and BMP signaling, respectively) with a common mediator SMAD (co-SMAD; SMAD4). Using PLA targeting different subunits of the trimeric SMAD-complex, i.e., either R-SMAD/co-SMAD or R-SMAD/R-SMAD, the formation of active SMAD transcription factor complexes can be measured quantitatively and spatially using fluorescence microscopy.
The usage of flow slides with 6 small parallel channels, that can be connected in series, allows for the investigation of the transcription factor complex formation and inclusion of necessary controls.
The workflow explained here can be easily adapted for studies targeting the proximity of SMADs to other transcription factors or to transcription factor complexes other than SMADs, in different fluid SS conditions. The workflow presented here shows a quick and effective way to study the fluid SS induced TGFβ/BMP signaling in ECs, both quantitatively and spatially.
Introduction
Proteins of the transforming growth factors beta (TGFβ) superfamily are pleiotropic cytokines with a variety of members, including TGFβs, bone morphogenetic proteins (BMPs), and Activins1,2. Ligand binding induces the formation of receptor oligomers leading to the phosphorylation and, thereby, activation of cytosolic regulatory SMAD (R-SMAD). Depending on the sub-family of ligands, different R-SMADs are activated1,2. While TGFβs and Activins mainly induce phosphorylation of SMAD2/3, BMPs induce SMAD1/5/8 phosphorylation. However, there are accumulating evidences that BMPs and TGFβs/Activins also activate R-SMADs of the respective other sub-family, in a process termed as 'lateral signaling'3,4,5,6,7,8 and that there are mixed SMAD complexes consisting of both, SMAD1/5 and SMAD2/3, members3,9. Two activated R-SMADs subsequently form trimeric complexes with the common mediator SMAD4. These transcription factor complexes are then able to translocate into the nucleus and regulate the transcription of target genes. SMADs can interact with a variety of different transcriptional co-activators and co-repressors, leading to the diversification of the possibilities to regulate target genes10. Deregulation of SMAD signaling has severe implications in a variety of diseases. In line with this, unbalanced TGFβ/BMP signaling may lead to severe vascular pathologies, such as pulmonary artery hypertension, hereditary hemorrhagic telangiectasia, or atherosclerosis3,11,12,13,14.
Endothelial cells (ECs) form the innermost layer of blood vessels and are, therefore, exposed to shear stress (SS), a frictional force exerted by the viscous flow of the blood. Interestingly, ECs residing at the parts of the vasculature, which are exposed to high levels of uniform, laminar SS, are kept in a homeostatic and quiescent state. In contrast, ECs that experience low, non-uniform SS, e.g., at bifurcations or the lesser curvature of the aortic arch, are proliferative and activate inflammatory pathways15. In turn, sites of dysfunctional ECs are prone to develop atherosclerosis. Interestingly, ECs in these atheroprone areas display aberrantly high levels of activated SMAD2/3 and SMAD1/516,17,18. In this context, enhanced TGFβ/BMP signaling was found to be an early event in the development of atherosclerotic lesions19 and interference with BMP signaling was found to markedly reduce vascular inflammation, atheroma formation, and associated calcification20.
Proximity Ligation Assay (PLA) is a biochemical technique to study protein-protein interactions in situ21,22. It relies on the specificity of antibodies of different species that can bind target proteins of interest, allowing highly specific detection of endogenous protein interactions at a single-cell level. Here, primary antibodies have to bind to their target epitope at a distance of less than 40 nm to allow for the detection23. Therefore, PLA is greatly beneficial over traditional co-immunoprecipitation approaches, where several million cells are needed to detect endogenous protein interactions. In PLA, species-specific secondary antibodies, covalently linked to DNA fragments (termed Plus and Minus probes), bind the primary antibodies and if the proteins of interest interact, Plus and Minus probes come in close proximity. The DNA gets ligated in the following step and the rolling circle amplification of the circular DNA is made possible. During amplification, fluorescently labeled complementary oligonucleotides bind to the synthesized DNA, allowing these protein interactions to be visualized by conventional fluorescence microscopy.
The protocol described here enables scientists to quantitatively compare the number of active SMAD transcription complexes at atheroprotective and atheroprone SS conditions in vitro using PLA. SS is generated via a programmable pneumatic pump system that is able to generate laminar unidirectional flow of defined levels and allows stepwise increases of flow rates. This method allows for the detection of interactions between SMAD1/5 or SMAD2/3 with SMAD4, as well as mixed-R-SMAD complexes. It can easily be expanded to analyze interactions of SMADs with transcriptional co-regulators or to transcription factor complexes other than SMADs. Figure 1 shows the major steps of the protocol presented below.
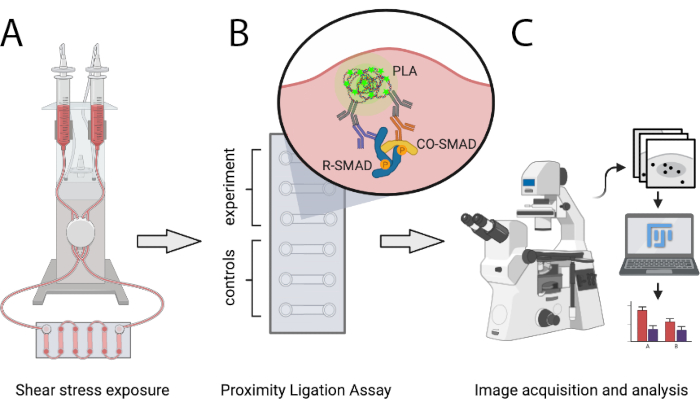
Figure 1: Schematic representation of the protocol described. (A) Cells seeded in 6-channel slides are exposed to shear stress with a pneumatic pump system. (B) Fixed cells are used for PLA experiment or for control conditions. (C) Images of PLA experiments are acquired with a fluorescence microscope and are analyzed using ImageJ analysis software. Please click here to view a larger version of this figure.
Protocol
1. Cell culture and fluid shear stress exposure
NOTE: Human umbilical vein ECs (HUVECs) were used as an example to study SS induced interaction of SMADs. The protocol described below can be applied to every SS responsive cell type.
- Coat 6-channel slide with 0.1% porcine skin gelatin in PBS for 30 min at 37 °C.
- Seed HUVECs in pre-coated 6-channel slides at a density of 2.5 x 106 cells per mL in 30 µL of M199 full medium.
NOTE: For further information on how to seed cells in the flow slide, see reference24. - Let cells adhere for 1 h at 37 °C in a humidified incubator.
- Add 60 µL of pre-warmed M199 full medium to each of the reservoirs.
- Culture for 2 days, with a gentle medium exchange once a day, at 37 °C in a humidified incubator.
- Aspirate the reservoirs completely, add 120 µL of pre-warmed M199 full medium in one of the reservoirs, and aspirate from the other side.
- Add 60 µL of pre-warmed M199 full medium to both reservoirs.
- Assemble and start the flow set-up as detailed in the reference25.
- Mount tubing on fluidic units. Here, silicone tubing with a diameter of 0.8 mm and 1.6 mm are used to apply shear stress of 1 dyn/cm2 and 30 dyn/cm2, respectively.
NOTE: The material and tubing length should remain constant, as changes could influence the resulting shear stress. In general, other combinations of pump systems and tubing can be used, as long as the resulting shear stress is known, and the pump creates a steady laminar flow. - Fill the reservoirs with an appropriate amount of pre-warmed M199 full medium (minimum 10 mL).
- Connect fluidic units with the tubing to the pump system and perform a pre-run without cells to equilibrate the medium and to remove any remaining air25.
- Serially connect the single channels on the 6-channel slide to one another by using serial connection tubing. The first and the last channel on the slide will be connected to the tubing assembled in 1.6.1 (see Figure 1A for a scheme). Be careful not to introduce any air into the system as this could severely harm the cells. Further information on the serial connection can be found in reference26.
- For the exposure of cells to high levels of shear stress (>20 dyn/cm2), use a ramp phase, i.e., increase the shear stress stepwise with adaptation phases. Steps can be set in increments of 5 dyn/cm2 per 30 min.
- Mount tubing on fluidic units. Here, silicone tubing with a diameter of 0.8 mm and 1.6 mm are used to apply shear stress of 1 dyn/cm2 and 30 dyn/cm2, respectively.
2. Fixation
- Detach slides from the pumps after the fluid SS exposure. Use clamps on the tubing when detaching, to avoid the medium spill in the incubator.
- Immediately transfer flow slides on ice, while the remaining tubing is detached sequentially. When removing the tubing from reservoirs, the reservoir on the other side should be kept closed with a finger to avoid trapping air bubbles in the channel. This might interfere with fixation steps.
- Keeping the cells on ice, aspirate the medium carefully from the reservoirs but not from the channel where the cells reside. Subsequently, wash samples with cold sterile PBS (4 °C) with three times the channel volume (90 µL). Add PBS in one reservoir and aspirate carefully from the other reservoir. Repeat this step in each of the 6 channels per slide.
NOTE: For all washing and incubation steps the respective solution is added in one of the reservoirs which leads to an exchange of solutions in the channel. To allow for complete substitution of solutions in the channel, the excess solution is then aspirated from the other reservoir. Solution on the top of the cells in the channel is not removed. Cells should not dry at any time. Therefore, it is important to wash carefully without any air bubble insertion into the slides. - Fix the cells by adding 90 µL of buffered 4% PFA solution in the same reservoir where the PBS was added beforehand and similarly aspirate the liquid from the other reservoir. Repeat this step in each channel in each slide. After the addition of PFA solution, transfer the samples from ice to room temperature (RT) and incubate for 20 min.
CAUTION: PFA is toxic and should be handled carefully. Use gloves and work under a fume hood. - Wash cells 3x with PBS (RT) by adding it in one reservoir and aspirating carefully from the other reservoir. Empty just the reservoirs, ensuring not to dry out the channel. Repeat this step for each of the 6 channels per slide.
- Quench the PFA-fixation by adding 90 µL of ambient 50 mM ammonium chloride in PBS in one of the reservoirs. Aspirate excess solution from the other reservoir. Repeat for each channel in the slide. Incubate the samples for 10 min at RT.
- Wash as described in step 2.5.
NOTE: At this point, the samples may be stored at 4 °C overnight, or the protocol can be immediately continued with blocking and primary antibody incubation (see step 3).
3. Blocking and primary antibody incubation
- To permeabilize the cells, add 90 µL of 0.3% Triton-X-100 in PBS in an emptied reservoir, and aspirate from the other reservoir for each channel. Incubate for 10 min at RT.
- Wash as described in step 2.5.
- Add 90 µL of sterile PLA blocking solution in one reservoir of a channel and aspirate from the other side. Repeat this step for each channel. Block for 1 h at 37 °C in a humidified chamber.
- To make a humidified chamber, use a 10 cm dish with wet tissue sealed with wax film and place the dish in the incubator. Alternatively, other humidity chamber formats can be used that supply a humid atmosphere.
NOTE: Alternatively, self-made blocking solution can be used (e.g., 3% (w/v) BSA in PBS, sterile filtered).
- To make a humidified chamber, use a 10 cm dish with wet tissue sealed with wax film and place the dish in the incubator. Alternatively, other humidity chamber formats can be used that supply a humid atmosphere.
- Prepare primary antibodies (1:100) in PLA antibody diluent. Prepare 30 µL of the solution per channel. Add both primary antibodies simultaneously and vortex.
NOTE: Alternatively, self-made antibody diluent can be used (e.g., 1% (w/v) BSA in PBS). Antibodies used here are combinations of SMAD1-SMAD2/3, SMAD2/3-SMAD4 and phospho-SMAD1/5-SMAD4. Detailed information can be found in the Table of Materials. - Before the application of primary antibodies, aspirate the blocking solution from the reservoirs and, also, carefully from the channel. Pipette 30 µL of the primary antibody solution immediately into the empty channel by tilting the channel while adding the solution.
NOTE: Perform the removal of the blocking solution and addition of the antibody solution channel-by-channel to ensure cells do not dry out in between. - Incubate samples with the primary antibodies overnight in humidified chambers at 4 °C.
NOTE: The incubation can also be performed for 1 h at room temperature, if interested in continuing with the following steps on the same day.
4. PLA probe incubation
NOTE: For all steps in section 4.1-7.3, the washing buffers A and B are stored at 4 °C and need to be warmed to RT prior to the use.
- Dilute PLA-probes (+)-mouse and (-)-rabbit to 1:5 in PLA antibody diluent (or 1% BSA) solution. Prepare 30 µL per channel.
- Wash samples 2x for 5 min using 90 µL of the wash buffer A at RT by adding it in one of the reservoirs and aspirating carefully from the other reservoir. Repeat this step for each of the 6 channels per slide.
- Aspirate the wash buffer A carefully and add 30 µL of PLA probe solution (prepared in step 4.1), similar to the addition of primary antibodies in step 3.5.
- Incubate samples for 1 h at 37 °C in a humidified chamber.
5. Ligation
- Wash samples 2x for 5 min using 90 µL of the wash buffer A at RT, as described in 4.2.
- Prepare a 1:5 dilution of the ligation buffer in deionized water. Use this buffer to dilute the ligase enzyme to 1:40 (on ice). Use 30 µL per channel.
- Aspirate the wash buffer A completely and add the ligation solution as described in 3.5.
- Incubate samples for 30 min at 37 °C in a humidified chamber.
6. Amplification
- Wash samples 2x for 2 min using 90 µL of wash buffer A at RT, as described in 4.2.
- Prepare the amplification buffer by diluting it 1:5 in deionized water and use it to dilute the polymerase enzyme to 1:80 (on ice). Protect from light. Prepare 30 µL per channel.
- Aspirate the wash buffer A completely and immediately add the prepared amplification solution into the empty channel, as described in 3.5. Incubate samples for 100 min at 37 °C in a humidified chamber.
7. Mounting
- Wash samples 2x for 10 min using 90 µL of Wash Buffer B at RT as described in 4.2. Add DAPI (1:500) from 1 mg/mL stock solution (in deionized water) in the first wash to stain nuclei. Do not dry the channel.
- Dilute the wash buffer B in deionized water (1:10) and wash 1x with 90 µL of 0.1x buffer B solution as described in 4.2.
- Aspirate the wash buffer B completely and immediately add 2-3 drops of the liquid mounting medium into one reservoir. Distribute it in the channel by tilting the slide. Store samples at 4 °C in a humidified environment until imaging.
8. Image acquisition
- Acquire images using a fluorescence microscope. Ensure that the respective filters fitting the fluorescent PLA probes are available.
NOTE: It is beneficial to make use of a confocal microscope, if possible, as obtained PLA spots are more defined. This also supports further image processing and data analysis.
9. Image analysis and quantification using ImageJ/FIJI
- Process exported images (.tiff) with an image processing program, such as ImageJ27.
NOTE: All scripts used within this study and that are necessary for the automatic counting of cellular, nuclear, and all PLA events (per cell) can be found in a GitHub repository: https://github.com/Habacef/Proximity-Ligation-Assay-analysis. Perform statistical analysis using any suitable program or tool.
Results
We have previously used PLA to detect interactions of different SMAD proteins3 and analyzed shear stress induced changes in SMAD phosphorylation28.
Here, both methods were combined with the protocol described above. HUVECs were subjected to shear stress of 1 dyn/cm2 and 30 dyn/cm2 and analyzed for interactions of SMAD transcription factors. We show that, when compared to the high shear stress (30 dyn/cm2), the low...
Discussion
The PLA based protocol described here offers an efficient way to determine close proximity of two proteins (e.g., their direct interaction) in ECs exposed to shear stress with quantitative and spatial resolution. By using flow slides with multiple parallel channels, several different protein interactions can be examined at the same time in cells under identical mechanical conditions. In contrast, custom-build flow chamber systems often make use of a single channel that is built around a glass coverslip, which would allow...
Disclosures
The authors declare no conflict of interest.
Acknowledgements
We thank Dr. Maria Reichenbach and Dr. Christian Hiepen for their support on the flow-set up system and Eleanor Fox and Yunyun Xiao for critically reading the manuscript. P-L.M. was funded by the international Max Planck Research School IMPRS-Biology and Computation (IMPRS-BAC). PK received funding by the DFG-SFB1444. Figure 1 was created using BioRender.
Materials
| Name | Company | Catalog Number | Comments |
| µ-Slide VI 0.4 | ibidi | 80606 | 6-channel slide |
| Ammonium Chloride | Carl Roth | K298.1 | Quenching |
| Bovine Serum Albumin | Carl Roth | 8076.4 | Blocking |
| DAPI | Sigma Aldrich/ Merck | D9542 | Stain DNA/Nuclei |
| DPBS | PAN Biotech | P04-53500 | PBS |
| Duolink In Situ Detection Reagents Green | Sigma Aldrich/ Merck | DUO92014 | PLA kit containing Ligase, ligation buffer, polymerase and amplification buffer (with green labeled oligonucleotides) |
| Duolink In Situ PLA Probe Anti-Mouse MINUS | Sigma Aldrich/ Merck | DUO92004 | MINUS probe |
| Duolink In Situ PLA Probe Anti-Rabbit PLUS | Sigma Aldrich/ Merck | DUO92002 | PLUS probe |
| Duolink In Situ Wash Buffers, Fluorescence | Sigma Aldrich/ Merck | DUO82049 | PLA wash buffers A and B |
| Endothelial Cell Growth Supplement | Corning | supplement for medium (ECGS) | |
| Fetal calf Serum | supplement for medium | ||
| FIJI | Image Analysis software | ||
| Formaldehyde solution 4% buffered | KLINIPATH/VWR | VWRK4186.BO1 | PFA |
| Full medium | M199 basal medium +20 % FCS +1 % P/S + 2 nM L-Glu + 25 µg/mL Hep + 50 µg/mL ECGS | ||
| Gelatin from porcine skin, Type A | Sigma Aldrich | G2500 | Use 0.1% in PBS for coating of flow channels |
| GraphPad Prism v.7 | GarphPad | Statistical Program used for the Plots and statistical calculations | |
| Heparin sodium salt from porcine intestinal mucosa | Sigma Aldrich | H4784-250MG | supplement for medium (Hep) |
| HUVECs | |||
| ibidi Mounting Medium | ibidi | 50001 | Liquid mounting medium |
| ibidi Pump System | ibidi | 10902 | pneumatic pump |
| Leica TCS SP8 | Leica | confocal microscope | |
| L-Glutamin 200mM | PAN Biotech | P04-80100 | supplement for medium (L-Glu) |
| Medium 199 | Sigma Aldrich | M2154 | Base medium |
| mouse anti- SMAD1 Antibody | Abcam | ab53745 | Suited for PLA |
| mouse anti- SMAD2/3 Antibody | BD Bioscience | 610843 | Not suited for PLA in combination with CST 9515 |
| mousee anti- SMAD4 Antibody | Sanata Cruz Biotechnology | sc-7966 | Suited for PLA |
| Penicillin 10.000U/ml /Streptomycin 10mg/ml | PAN Biotech | P06-07100 | supplement for medium (P/S) |
| Perfusion Set WHITE | ibidi | 10963 | Tubings used for 1 dyn/cm2 |
| Perfusion Set YELLOW and GREEN | ibidi | 10964 | Tubings used for 30 dyn/cm2 |
| rabbit anti- phospho SMAD1/5 Antibody | Cell Signaling Technologies | 9516 | Suited for PLA |
| rabbit anti- SMAD2/3 XP Antibody | Cell Signaling Technologies | 8685 | Suited for PLA |
| rabbit anti- SMAD4 Antibody | Cell Signaling Technologies | 9515 | Not suited for PLA in combination with BD 610843 |
| Serial Connector for µ-Slides | ibidi | 10830 | serial connection tubes |
| Triton X-100 | Carl Roth | 6683.1 | Permeabilization |
References
- Yadin, D., Knaus, P., Mueller, T. D. Structural insights into BMP receptors: Specificity, activation and inhibition. Cytokine and Growth Factor Reviews. 27, 13-34 (2016).
- Sieber, C., Kopf, J., Hiepen, C., Knaus, P. Recent advances in BMP receptor signaling. Cytokine and Growth Factor Reviews. 20 (5-6), 343-355 (2009).
- Hiepen, C., et al. BMPR2 acts as a gatekeeper to protect endothelial cells from increased TGFβ responses and altered cell mechanics. PLoS Biology. 17 (12), 3000557 (2019).
- Hildebrandt, S., et al. ActivinA induced SMAD1/5 Signaling in an iPSC derived EC model of Fibrodysplasia Ossificans Progressiva (FOP) can be rescued by the drug candidate saracatinib. Stem Cell Reviews and Reports. , (2021).
- Goumans, M. J., et al. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. The EMBO Journal. 21 (7), 1743-1753 (2002).
- Goumans, M. J., et al. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFbeta/ALK5 signaling. Molecular Cell. 12 (4), 817-828 (2003).
- Daly, A. C., Randall, R. A., Hill, C. S. Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Molecular and Cellular Biology. 28 (22), 6889-6902 (2008).
- Ramachandran, A., et al. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife. 7, 31756 (2018).
- Flanders, K. C., et al. Brightfield proximity ligation assay reveals both canonical and mixed transforming growth factor-β/bone morphogenetic protein Smad signaling complexes in tissue sections. The Journal of Histochemistry and Cytochemistry : The Official Journal of The Histochemistry Society. 62 (12), 846-863 (2014).
- Miyazono, K., Maeda, S., Imamura, T., Dijke, P. T., Heldin, C. -. H. . Smad Signal Transduction: Smads in Proliferation, Differentiation and Disease. , 277-293 (2006).
- Goumans, M. J., Zwijsen, A., Ten Dijke, P., Bailly, S. Bone morphogenetic proteins in vascular homeostasis and disease. Cold Spring Harbor Perspectives in Biology. 10 (2), 031989 (2018).
- Cai, J., Pardali, E., Sánchez-Duffhues, G., ten Dijke, P. BMP signaling in vascular diseases. FEBS Letters. 586 (14), 1993-2002 (2012).
- Cunha, S. I., Magnusson, P. U., Dejana, E., Lampugnani, M. G. Deregulated TGF-β/BMP signaling in vascular malformations. Circulation research. 121 (8), 981-999 (2017).
- MacCarrick, G., et al. Loeys-Dietz syndrome: a primer for diagnosis and management. Genetics in Medicine : An Official Journal of the American College of Medical Genetics. 16 (8), 576-587 (2014).
- Baeyens, N., Bandyopadhyay, C., Coon, B. G., Yun, S., Schwartz, M. A. Endothelial fluid shear stress sensing in vascular health and disease. The Journal of Clinical Investigation. 126 (3), 821-828 (2016).
- Min, E., et al. Activation of Smad 2/3 signaling by low shear stress mediates artery inward remodeling. bioRxiv. , 691980 (2019).
- Zhou, J., et al. BMP receptor-integrin interaction mediates responses of vascular endothelial Smad1/5 and proliferation to disturbed flow. Journal of Thrombosis and Haemostasis. 11 (4), 741-755 (2013).
- Zhou, J., et al. Force-specific activation of Smad1/5 regulates vascular endothelial cell cycle progression in response to disturbed flow. Proceedings of the National Academy of Sciences of the United States of America. 109 (20), 7770-7775 (2012).
- van Dijk, R. A., et al. Visualizing TGF-β and BMP signaling in human atherosclerosis: A histological evaluation based on Smad activation. Histology and Histopathology. 27 (3), 387-396 (2012).
- Derwall, M., et al. Inhibition of bone morphogenetic protein signaling reduces vascular calcification and atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 32 (3), 613-622 (2012).
- Fredriksson, S., et al. Protein detection using proximity-dependent DNA ligation assays. Nature Biotechnology. 20 (5), 473-477 (2002).
- Söderberg, O., et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature Methods. 3 (12), 995-1000 (2006).
- Alam, M. S. Proximity Ligation Assay (PLA). Current Protocols in Immunology. 123 (1), 58 (2018).
- Application Note 03: Growing Cells in µ-Channels. ibidi Available from: https://ibidi.com/img/cms/support/AN/AN03_Growing_cells.pdf (2012)
- Application Note 13: HUVECs under perfusion. ibidi Available from: https://ibidi.com/img/cms/support/AN/AN13_HUVECs_under_perfusion.pdf (2019)
- ibidi. Application Note 31: Instructions µ-Slide VI 0.4. ibidi. , (2013).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Reichenbach, M., et al. Differential impact of fluid shear stress and YAP/TAZ on BMP/TGF-β induced osteogenic target genes. Advanced Biology. 5 (2), 2000051 (2021).
- Hiepen, C., Mendez, P. L., Knaus, P. It takes two to tango: Endothelial TGFβ/BMP signaling crosstalk with mechanobiology. Cells. 9 (9), 1965 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved