Method Article
Production of siRNA-Loaded Lipid Nanoparticles using a Microfluidic Device
In This Article
Summary
Microfluidic-based lipid nanoparticle (LNP) production methods have attracted attention in drug delivery systems (DDSs), including RNA delivery. This protocol describes the fabrication, LNP (siRNA-loaded LNP) production, and LNP evaluation processes using our original microfluidic device named iLiNP.
Abstract
The development of functional lipid nanoparticles (LNPs) is one of the major challenges in the field of drug delivery systems (DDS). Recently, LNP-based RNA delivery systems, namely, RNA-loaded LNPs have attracted attention for RNA therapy. In particular, mRNA-loaded LNP vaccines were approved to prevent COVID-19, thereby leading to the paradigm shift toward the development of next-generation nanomedicines. For the LNP-based nanomedicines, the LNP size is a significant factor in controlling the LNP biodistribution and LNP performance. Therefore, a precise LNP size control technique is indispensable for the LNP production process. Here, we report a protocol for size controlled LNP production using a microfluidic device, named iLiNP. siRNA loaded LNPs are also produced using the iLiNP device and evaluated by in vitro experiment. Representative results are shown for the LNP size, including siRNA-loaded LNPs, Z-potential, siRNA encapsulation efficiency, cytotoxicity, and target gene silencing activity.
Introduction
Lipid nanoparticle (LNP) is one of the most widely used nanocarriers for RNA delivery systems. Recently, mRNA-loaded LNPs have been approved as vaccines for the prevention of COVID-191,2,3. Generally, the size of LNP plays a crucial role in the biodistribution and drug delivery systems (DDS) performance, including gene silencing or protein expression4,5,6. Therefore, a precise LNP size control method is required for the LNP production process.
For the production of size controlled LNPs, microfluidic devices have attracted attention over the years7. In 2018, the first Food and Drug Administration (FDA)-approved siRNA-loaded LNPs (e.g., Onpattro) was developed using the microfluidic device8,9. In the microfluidic-based LNP production method, a lipid solution and an aqueous solution are introduced separately into the microfluidic device, and then mixed in the microchannel. To enhance the mixing efficiency, the chaotic mixer device has been used for the LNP production10,11,12. The chaotic mixer device makes it possible to produce specific-sized LNPs.
A simple microfluidic device, named invasive lipid nanoparticle production (iLiNP), equipped with baffle structures, has been developed to control the LNP size precisely13,14. In comparison with the chaotic mixer device, the iLiNP device was able to control the LNP size ranged from 20 to 100 nm at 10 nm intervals. In addition, the iLiNP device produced siRNA-loaded LNPs6, mRNA-loaded LNPs15, ribonucleoprotein-loaded LNPs16, and exosome-like LNPs17. The aim of this paper is to introduce the fabrication andsiRNA-loaded LNP production process of the iLiNP device and describe the LNP evaluation process produced by the iLiNP device.
Protocol
1. Fabrication of the iLiNP device
NOTE: The iLiNP device is fabricated using the standard soft lithography method18. The detailed fabrication protocol was reported previously10,13.
- SU-8 mold fabrication
- Pour SU-8 3050 onto a 3-inch silicon wafer. Spin coat the silicon wafer to obtain a 100 µm thick SU-8 layer.
- Bake the silicon wafer by placing on a hotplate at 65 °C for 5 min and 95 °C for 45 min.
- After baking, place the silicon wafer on the stage of a desktop maskless lithography system.
- Expose the silicon wafer to UV light at 365 nm for 1.5 s per one position.
NOTE: A desktop maskless lithography system was used in this experiment. The system automatically exposes UV light at a divided irradiation area (one position) of the microchannel. - After UV irradiation, bake the silicon wafer on the hotplate at 65 °C for 1 min and 95 °C for 5 min.
- Cool the silicon wafer, and then soak in a SU-8 developer for 15 min to remove unexposed SU-8.
- Treat the SU-8 mold withvapor of Trichloro(1H,1H,2H,2H-perfluorooctyl)silane using a desiccator and vacuum pump.
- Fabrication of the iLiNP device
- Mix the silicone base and polydimethylsiloxane (PDMS) curing agent in a 10:1 ratio (w/w).
- Degas the mixture using a vacuum pump and a desiccator.
NOTE: PDMS was degassed using a vacuum pump for 10 min at room temperature. - Pour the degassed PDMS onto the SU-8 mold in a 100 mm Petri dish up to 0.5 to 1 cm thickness, followed by baking in an oven at 80 °C for 1 h.
- Cool the mold, and then peel the PDMS substrate from the SU-8 mold using a tweezer.
- Punch three holes (0.5 mm) in the PDMS substrate. Bond the PDMS substrate and a glass slide using an oxygen plasma cleaner to build an iLiNP device (see Figure 1)13.
- Connect three PEEK capillaries (I.D. 0.3 mm, O.D. 0.5 mm) to the inlets and outlet of the iLiNP device and cure with a superglue.
NOTE: The length of the PEEK capillaries is adjustable and depends on the experiment.
2. Preparation of lipid solutions
- Prepare lipid/ethanol solutions: 13.4 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 10 mM 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC), 20 mM 1,2-dioleoyloxy-3-trimethylammonium propane (DOTAP), 5 mM 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2k), and 20 mM cholesterol. Store the stock solutions at -20 °C prior to the experiment.
- To produce the siRNA-loaded LNPs, mix DOTAP, DSPC, cholesterol, and DMG-PEG2k solutions at a molar ratio of 50/10/38.5/1.5. The total lipid concentration is adjusted to 8 mM.
3. Preparation of aqueous solutions
- Prepare aqueous solutions: 154 mM NaCl (saline), 25 mM acetate buffer at pH 4.0 using DNase/RNase-free distilled water.
- Filter the solutions through 0.2 µm sized membrane filters or syringe filters.
4. Preparation of the siRNA/buffer solution
- Dissolve 70 µg of siGL4 into 1 mL of 25 mM acetate buffer (pH 4.0).
NOTE: siGL4 is used for the knockdown of luciferase gene.
5. Set up of the iLiNP device and production of LNPs
NOTE: See Figure 1 for the schematics.
- Fill 1 mL glass syringes with lipid and aqueous solutions (from steps 3.1 and 4.1 in individual syringes), respectively.
NOTE: Adjust the lipid and aqueous solution volume depending on the amount required for the LNP evaluation experiment. - Connect the glass syringes to the PEEK capillaries using syringe connectors.
- Set the flow rate of the lipid and aqueous solutions.
NOTE: The flow rate ratio (FRR) of the aqueous phase to the lipid phase is ranged from 3:1 to 9:1. - Introduce the lipid and aqueous solutions separately into the iLiNP device using syringe pumps.
- Collect LNP suspensions in a microtube from the outlet of the iLiNP device (Figure 1).
6. Dialysis of the LNP suspension and LNP size measurement
- Dialyze the LNP suspension using a dialysis membrane (12−14 kDa MW cutoffs) at 4 °C overnight against saline or D-PBS for POPC LNPs and siRNA-loaded LNPs, respectively.
NOTE: POPC is not dissolved into saline (Please see 2.1). POPC/ethanol solution is diluted with saline in the iLiNP device. - Collect the dialyzed LNP suspensions in microtubes.
- Pipette 20-30 µL of the LNP suspension to a micro quartz cell.
- Measure the LNP size, LNP size distribution, and polydispersity index by dynamic light scattering (DLS).
7. Measurement of Z-potential of the LNP
NOTE: For the measurement of Z-potential, a particle analyzer (see Table of Materials) was used following the manufacturer's instruction.
- Dilute the LNP suspension obtained from step 6.1, 35 times with 10 mM HEPES buffer (pH 7.4).
- Pipette 700 to 1000 µL of the diluted LNP suspension to a capillary cell.
- Measure the Z-potential as per the manufacturer's instruction.
8. siRNA encapsulation efficiency by RiboGreen assay
NOTE: Ribogreen assay is performed to evaluate the siRNA encapsulation into LNPs19. Ribogreen assay can measure the amount of RNAs inside and outside of LNPs with/without a surfactant (e.g., TritonX-100).
- Dilute 2 mg/mL of siGL4 with 10 mM HEPES buffer (pH 7.4) to 500 ng/mL siGL4 solution.
- Prepare the dilution series (0, 12.5, 25, 50, 100, 200 ng/mL) of siGL4 solution to make a calibration curve for Triton (+) and Triton (-) samples.
- Dilute the LNP suspension 100 times with 10 mM HEPES buffer (pH 7.4).
- Mix the following for Triton (+) solution: 980 µL of 10 mM HEPES (pH 7.4), 20 µL of 10% w/v TritonX-100, and 1.25 µL of RiboGreen for 10 wells of a 96-well microplate.
- Mix the following for Triton (-) solution: 1000 µL of 10 mM HEPES (pH 7.4) and 1.25 µL of RiboGreen for 10 wells of a 96-well microplate.
- Pipette 100 µL of the dilution series of siGL4 solution and diluted LNP suspensions into the wells of a black 96-well microplate.
NOTE: The dilution series of siGL4 solution and diluted LNP suspensions were dispensed into four microwells per condition. - Pipette 100 µL of the detection solution (TritonX-100 (+) or Triton (-)) into the wells.
NOTE: The detection solution (TritonX-100 (+)) was dispensed into two wells per sample per condition, and the TritonX-100 (-) solution was dispensed into the remaining two wells per sample condition. - Incubate the microplate for 5 min at room temperature.
- Measure the fluorescence intensity using a microplate readerat a wavelength of 475 nm.
- Calculate the siRNA encapsulation efficiency from the following equation19.
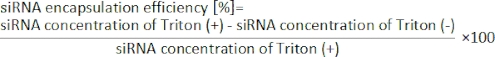
9. Cell culture
- Prepare a growth medium containing DMEM, heat-inactivated 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 400 µg/mL G418.
- Culture HeLa cells stably expressing firefly and Renilla luciferase (HeLa-dluc) in a 100 mm TC-treated cell culture dish containing the growth medium at 37 °C in a 5% CO2 incubator.
10. Cell viability assay
- Seed 100 µL of a suspension of HeLa cells in the growth medium (6 x 103 cells/well) in a 96-well microplate.
NOTE: Cells were counted using a cell counter plate and a microscope. - Incubate the microplate for 24 h at 37 °C in a 5% CO2 incubator.
- Dilute the siRNA-loaded LNPs with DMEM (FBS (-)) at the concentrations of 10 and 100 nM siRNAs.
- Dispense 100 µL of the diluted siRNA-loaded LNP suspension per well.
- Incubate the microplate for 4 h at 37 °C in a 5% CO2 incubator.
- Remove the LNP suspension and add 100 µL of DMEM (FBS (+)).
- Incubate the microplate for 20 h at 37 °C in a 5% CO2 incubator.
- Measure the cell viability using a commercially available kit according to the manufacturer's protocol.
NOTE: D-PBS (-) was used as the negative control.
11. Luciferase gene knockdown assay
- Seed 75 µL of a suspension of HeLa cells in the growth medium (4.5 x 103 cells/well) in a 96-well microplate.
- Incubate the microplate for 24 h at 37 °C in a 5% CO2 incubator.
- Dilute the siRNA-loaded LNPs with DMEM (FBS (-)) at the concentrations of 10 and 100 nM siRNAs.
- Dispense 75 µL of the diluted siRNA-loaded LNP suspension per one well.
- Incubate the microplate for 4 h at 37 °C in a 5% CO2 incubator.
- Remove the LNP suspension and add 75 µL of DMEM (FBS (+)).
- Incubate the microplate for 20 h at 37 °C in a 5% CO2 incubator.
- Measure the luciferase expression using a commercially available kit according to the manufacturer's protocol.
NOTE: We used D-PBS (-) as the negative control.
Results
Figure 2A,B shows the POPC LNP size distribution produced at different flow conditions. The microfluidic-based LNP preparation method can control the size of LNPs by the flow conditions such as the total flow rate (TFR) and the FRR. Compared with the typical microfluidic devices, including the chaotic mixer device and the flow-focusing microfluidic device, the iLiNP device enabled precise LNP size control ranging from 20 to 100 nm (Figure 2). Small-sized LNPs formed at high total flow rate conditions. In addition, the LNP sizes formed at the FRR of 5 were smaller than those of the FRR of 3, regardless of the total flow rate13.
siRNA-loaded LNPs were also prepared using the iLiNP device (Figure 3A). For the siRNA-loaded LNP preparation, DOTAP, a cationic lipid, was used to encapsulate the siRNA into the LNPs effectively. The iLiNP device produced 90 nm sized siRNA-loaded cationic LNPs with narrow distribution (Figure 3A,B). The siRNA encapsulation efficiency was 95% because of the electrostatic interaction between the cationic lipid and negatively charged siRNAs (Figure 3C).
Cytotoxicity and the gene silencing activity of 90 nm sized siRNA-loaded LNPs were evaluated as shown in Figure 4 and Figure 5. siRNA-loaded LNPs did show cytotoxicity at a dose of 10 and 100 nM siRNA. We also confirmed that the expression level of luciferase was decreased depending on the siRNA concentration. The siRNA-loaded LNPs suppressed 80% luciferase expression at a dose of 100 nM siRNA. The effect of LNP size on the gene silencing activity was reported previously6,13,17.
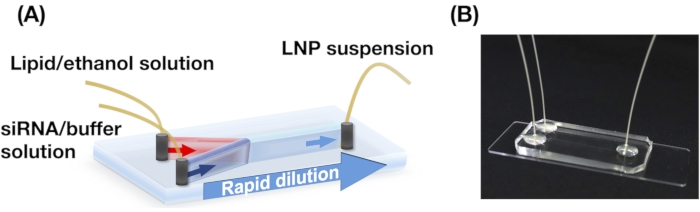
Figure 1: (A) Schematic illustration and (B) photograph of the iLiNP device. The iLiNP device comprises PDMS and glass substrates. The iLiNP device is connected to PEEK capillaries with a superglue. The lipid and siRNA/buffer solutions are separately introduced into the iLiNP device using syringe pumps. The LNP suspension is collected in a microtube. Please click here to view a larger version of this figure.
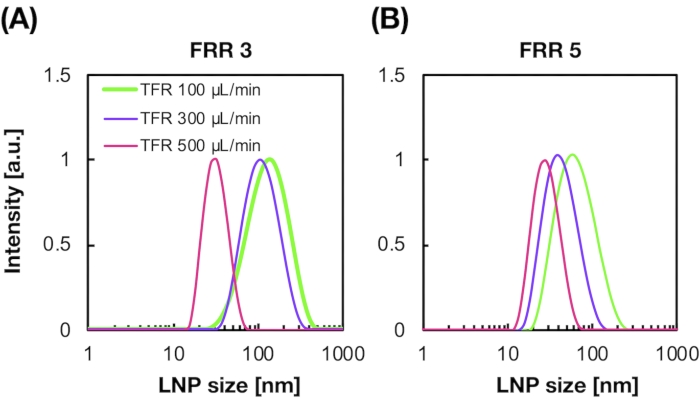
Figure 2: POPC LNP size distributions produced by the iLiNP device at the different flow rate ratios (FRR). The POPC LNP size is measured by dynamic light scattering (DLS). The POPC LNPs are prepared by changing the total flow rate and the FRR: (A) 3 FRR and (B) 5 FRR. Small-sized LNPs are formed at high total flow rate conditions. In addition, the LNP sizes formed at the FRR of 5 were smaller than those at the FRR of 3. Please click here to view a larger version of this figure.
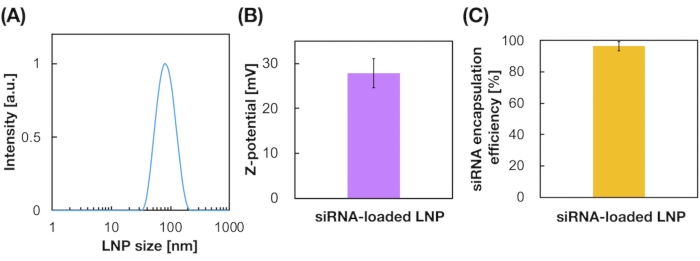
Figure 3: Characterization of the siRNA-loaded LNPs. (A) Size distribution of siRNA-loaded LNPs. siRNAs (siGL4) are encapsulated into the LNPs by electrostatic interaction between the cationic lipid (DOTAP) and negatively-charged siRNAs. (B) Z-potential of the siRNA-loaded LNPs. The LNP suspension was diluted with 10 mM HEPES buffer (pH 7.4) before the measurement. Data are represented as mean ± SD (Standard Deviation). n = 3. (C) siRNA encapsulation efficiency of the DOTAP-based LNPs. The encapsulation efficiency was determined by RiboGreen assay. Data are represented as mean ± SD. n = 3. Please click here to view a larger version of this figure.
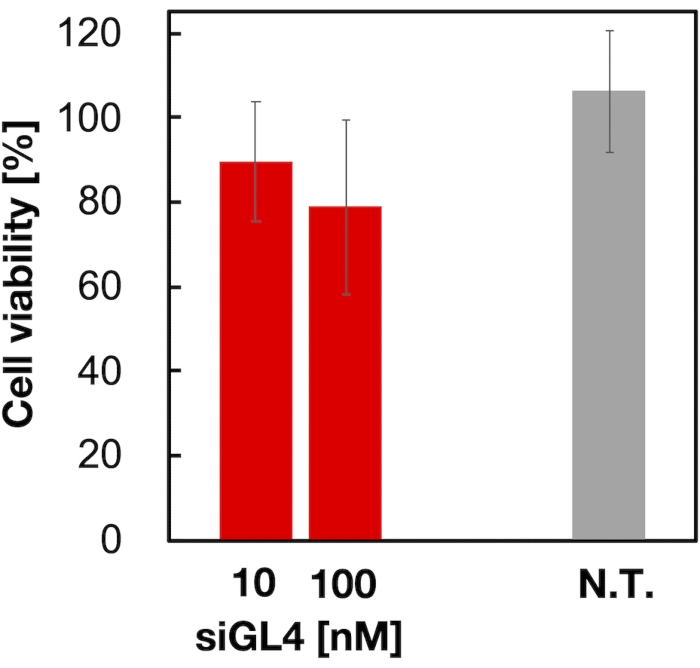
Figure 4: Cytotoxicity of the siRNA-loaded LNPs. siRNA-loaded LNPs were diluted with DMEM (FBS (-)) to obtain the siGL4 concentrations of 10 and 100 nM. The LNP suspensions are added to HeLa-dLuc cells and incubated for 4 h at 37 °C in a 5% CO2 incubator. N.T.: Non-treated (D-PBS(-)). Data are represented as the mean ± SD. n = 3. Please click here to view a larger version of this figure.
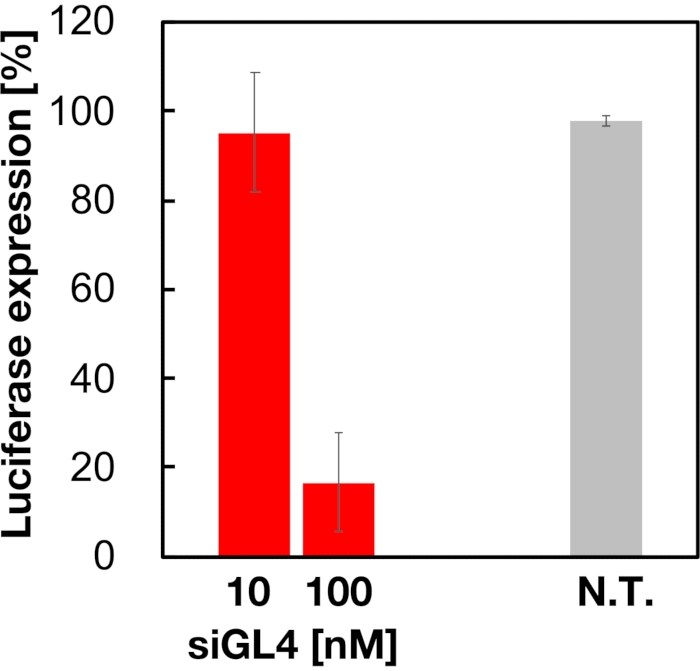
Figure 5: Luciferase gene knockdown activity treated with siRNA-loaded LNPs. siRNA-loaded LNPs are prepared in the same manner as cell viability assay. The luciferase expression level is measured using Dual-Glo Luciferase Assay System. N.T.: Non-treated (D-PBS(-)). Data are represented as mean ± SD. n = 3. Please click here to view a larger version of this figure.
Discussion
The LNP size affects the LNP biodistribution, anti-tumor effect, and gene silencing performance. Therefore, the LNP size control method is a significant technique for producing DDS nanomedicines, including RNA delivery systems. The aim of this paper is to introduce the iLiNP device for precise size tuning of LNPs and its application to the siRNA-loaded LNPs production. The iLiNP device was able to control the LNP size ranged from 20 to 100 nm (Figure 2)13. When the flow conditions, such as the total flow rate and the FRR are changed to control the LNP size, the LNP suspension should be collected after about 5 to 10 s to stabilize the solution flow. The LNP suspension collected from the outlet of the iLiNP device was dialyzed immediately against the buffer solution to remove ethanol and prevent LNP aggregation.
The LNP size control is one of the major challenges in the field of DDS. Generally, the conventional LNP production process, such as the lipid film hydration method, needs a size tuning process after the LNP production20. On the other hand, the microfluidic-based LNPs production method can produce the size-controlled LNPs by introducing the lipid and aqueous solutions into the microfluidic device6,11,13. Although the dialysis process is required to remove ethanol from the LNP suspension, a continuous process by the microfluidic device coupled with the tangential flow system promises the automation of the LNP production process14. According to the literature, the POPC LNP sizes were 50-60 nm and 30-60 nm, for the flow-focusing microfluidic device21 and the chaotic mixer device, respectively10. Compared with other microfluidic devices, the iLiNP device enables the POPC LNP size control in a wide range from 20 to 100 nm.
The fabrication process of the iLiNP device employed was the standard soft lithography. Thus, the iLiNP device can be mass-produced by rapid prototyping technique and prevent cross-contamination of solutions by using a disposable device. The iLiNP device can produce siRNA-loaded LNPs in the same manner as the POPC LNP production method. For the LNP production method using the iLiNP device, the user does not require any complicated procedures. For these reasons, the microfluidic-based LNP production method, including the iLiNP device, will be expected to be employed as the standard LNP production method. The protocol of this paper can be adapted to other microfluidic devices for LNP production. In addition, the production of mRNA-loaded LNPs is also enabled by changing the siRNA/buffer solution to a buffer solution containing mRNAs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by JST, CREST Grant Number JPMJCR17H1, Japan, JST, PRESTO Grant Number JPMJPR19K8, Japan, JST, SCORE, Japan, the Special Education and Research Expenses from the Ministry of Education, Culture, Sports, Science and Technology, JSPS KAKENHI Grant Number JP19KK0140, and Iketani Science and Technology Foundation.
Materials
| Name | Company | Catalog Number | Comments |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | NOF Corp. | MC-6081 | |
| 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2K) | NOF Corp. | GM-020 | |
| 1,2-dioleoyloxy-3-trimethylammonium propane (DOTAP) | NOF Corp. | CL-8181TA | |
| 1,2-Distearoyl-sn-glycero-3-phosphocholinev (DSPC) | NOF Corp. | MC-8080 | |
| 10 x D-PBS (-) | FUJIFILM Wako Pure Chemical Corp. | 048-29805 | |
| Acetic acid | FUJIFILM Wako Pure Chemical Corp. | 017-00251 | |
| CellTiter-Blue Cell Viability Assay | Promega | G8081 | |
| cholesterol | Sigma-Aldrich | C8667-5G | |
| Desktop maskless lithography system | NEOARK CORPORATION | DDB-701-DL4 | |
| Dialysis membrane | Repligen | 132697 | |
| Dual-Glo Luciferase Assay System | Promega | E2940 | |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | Lot: 42G6587K | |
| G418 | Nacalai Tesque | 08973-14 | |
| Glass substrate | Matsunami Glass Ind., Ltd. | S1111 | |
| Glass syringe | Hamilton | GASSTIGHT 1002 | |
| HeLa cell | HeLa-dluc cells were provided from Dr. Yusuke Sato at Hokkaido University | ||
| HEPES | FUJIFILM Wako Pure Chemical Corp. | 342-01375 | |
| Low-glucose Dulbecco’s modified Eagle medium (DMEM) | Sigma-Aldrich | D6046-500ML | |
| Oxygen plasma cleaner | Femto Science | CUTE-1MP/R | |
| Penicillin–streptomycin, trypsin (2.5%) | Thermo Fisher Scientific | 15140122 | |
| Quant-iT RiboGreen RNA Reagent | Thermo Fisher Scientific | R11491 | |
| siGL4 | Hokkaido System Science Co., Ltd | The sense and antisense strand sequences of siGL4 are 5'-CCGUCGUAUUCGUGAGCAATsT -3' and 5'-UUGCUCACGAAUACGACGGTsT -3', respectively. | |
| Silicon wafer | GTC | ||
| SILPOT 184 W/C (PDMS) | Dow Corning Toray Co., Ltd. | silicone base and curing agent are included | |
| Sodium acetate | FUJIFILM Wako Pure Chemical Corp. | 192-01075 | |
| Sodium chloride | FUJIFILM Wako Pure Chemical Corp. | 191-01665 | |
| SU-8 3050 | Nippon Kyaku Co., Ltd. | ||
| Syringe connector | Institute of microchemical Technology Co., Ltd. | ISC-011 | |
| Syringe pump | Chemyx | CX07200 | |
| trichloro(1H,1H,2H,2H-perfluorooctyl)silane | Sigma-Aldrich | 448931-10G | |
| TritonX-100 | Nacalai Tesque | 35501-15 | |
| UltraPure DNase/RNase-Free Distilled Water | Thermo Fisher Scientific | 10977015 | |
| Zetasizer Nano ZS | Malvern Instruments | ZEN3600 |
References
- Schoenmaker, L., et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. International Journal of Pharmaceutics. 601, 120586 (2021).
- Chung, Y. H., Beiss, V., Fiering, S. N., Steinmetz, N. F. COVID-19 Vaccine frontrunners and their nanotechnology design. ACS Nano. 14 (10), 12522-12537 (2020).
- Dong, Y., et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduction and Targeted Therapy. 5 (1), 237 (2020).
- Cabral, H., et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nature Nanotechnology. 6 (12), 815-823 (2011).
- Sato, Y., et al. Elucidation of the physicochemical properties and potency of siRNA-loaded small-sized lipid nanoparticles for siRNA delivery. Journal of Controlled Release. 229, 48-57 (2016).
- Kimura, N., et al. Three-dimensional, symmetrically assembled microfluidic device for lipid nanoparticle production. RSC Advances. 11 (3), 1430-1439 (2021).
- Maeki, M., Kimura, N., Sato, Y., Harashima, H., Tokeshi, M. Advances in microfluidics for lipid nanoparticles and extracellular vesicles and applications in drug delivery systems. Advanced Drug Delivery Reviews. 128, 84-100 (2018).
- Akinc, A., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nature Nanotechnology. 14 (12), 1084-1087 (2019).
- Kulkarni, J. A., Witzigmann, D., Chen, S., Cullis, P. R., vander Meel, R. Lipid nanoparticle technology for clinical translation of siRNA therapeutics. Accounts of Chemical Research. 52 (9), 2435-2444 (2019).
- Maeki, M., et al. Understanding the formation mechanism of lipid nanoparticles in microfluidic devices with chaotic micromixers. PLoS One. 12 (11), 0187962 (2017).
- Maeki, M., et al. A strategy for synthesis of lipid nanoparticles using microfluidic devices with a mixer structure. RSC Advances. 5 (57), 46181-46185 (2015).
- Belliveau, N. M., et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Molecular Therapy - Nucleic Acids. 1, 37 (2012).
- Kimura, N., et al. Development of the iLiNP Device: Fine Tuning the Lipid Nanoparticle Size within 10 nm for Drug Delivery. ACS Omega. 3 (5), 5044-5051 (2018).
- Kimura, N., et al. Development of a microfluidic-based post-treatment process for size-controlled lipid nanoparticles and application to siRNA delivery. ACS Applied Materials & Interfaces. 12 (30), 34011-34020 (2020).
- Hashiba, A., et al. The use of design of experiments with multiple responses to determine optimal formulations for in vivo hepatic mRNA delivery. Journal of Controlled Release. 327, 467-476 (2020).
- Suzuki, Y., et al. Lipid nanoparticles loaded with ribonucleoprotein-oligonucleotide complexes synthesized using a microfluidic device exhibit robust genome editing and hepatitis B virus inhibition. Journal of Controlled Release. 330, 61-71 (2020).
- Kimura, N., Maeki, M., Ishida, A., Tani, H., Tokeshi, M. One-step production using a microfluidic device of highly biocompatible size-controlled noncationic exosome-like nanoparticles for RNA delivery. ACS Applied Bio Materials. 4 (2), 1783-1793 (2021).
- Deng, T., Wu, H., Brittain, S. T., Whitesides, G. M. Prototyping of masks, masters, and stamps/molds for soft lithography using an office printer and photographic reduction. Analytical Chemistry. 72 (14), 3176-3180 (2000).
- Sato, Y., et al. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. Journal of Controlled Release. 163 (3), 267-276 (2012).
- Ong, S. G., Chitneni, M., Lee, K. S., Ming, L. C., Yuen, K. H. Evaluation of extrusion technique for nanosizing liposomes. Pharmaceutics. 8 (4), (2016).
- Mijajlovic, M., Wright, D., Zivkovic, V., Bi, J. X., Biggs, M. J. Microfluidic hydrodynamic focusing based synthesis of POPC liposomes for model biological systems. Colloids and Surfaces B: Biointerfaces. 104, 276-281 (2013).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved