A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Isolation and Characterization of the Natural Microbiota of the Model Nematode Caenorhabditis elegans
In This Article
Summary
Caenorhabditis elegans is one of the main model species in biology, yet almost all research is performed in the absence of its naturally associated microbes. The methods described here will help to improve our understanding of the diversity of associated microbes as a basis for future functional C. elegans research.
Abstract
The nematode Caenorhabditis elegans interacts with a large diversity of microorganisms in nature. In general, C. elegans is commonly found in rotten plant matter, especially rotten fruits like apples or on compost heaps. It is also associated with certain invertebrate hosts such as slugs and woodlice. These habitats are rich in microbes, which serve as food for C. elegans and which can also persistently colonize the nematode gut. To date, the exact diversity and consistency of the native C. elegans microbiota across habitats and geographic locations is not fully understood. Here, we describe a suitable approach for isolating C. elegans from nature and characterizing the microbiota of worms. Nematodes can be easily isolated from compost material, rotting apples, slugs, or attracted by placing apples on compost heaps. The prime time for finding C. elegans in the Northern Hemisphere is from September until November. Worms can be washed out of collected substrate material by immersing the substrate in buffer solution, followed by the collection of nematodes and their transfer onto nematode growth medium or PCR buffer for subsequent analysis. We further illustrate how the samples can be used to isolate and purify the worm-associated microorganisms and to process worms for 16S ribosomal RNA analysis of microbiota community composition. Overall, the described methods may stimulate new research on the characterization of the C. elegans microbiota across habitats and geographic locations, thereby helping to obtain a comprehensive understanding of the diversity and stability of the nematode's microbiota as a basis for future functional research.
Introduction
In nature, C. elegans is commonly found in rotten plant matter, especially rotten fruits like apples or on compost heaps1. It is also associated with certain invertebrate hosts such as slugs and woodlice2,3. These habitats are rich in microbes, which not only serve as food for the worm, but may also form stable associations with it. Information on the diversity of naturally associated microorganisms was only published in 20164,5,6. Since then, these and only a few more recent studies have revealed that C. elegans is associated with a variety of bacteria and fungi, most commonly including bacteria of the genus Pseudomonas, Enterobacter, Ochrobactrum, Erwinia, Comamonas, Gluconobacter, and several others6,7,8. Several associated bacteria can stably colonize the worm gut, although not all6,9,10,11,12. They are likely to be of key importance for our understanding of C. elegans biology because they can provide nutrition, protect against pathogens and possibly other stressors, and affect central life-history traits such as reproductive rate, development, or behavioral responses.
As an example, naturally associated isolates of the genera Pseudomonas, Ochrobactrum, and also Enterobacter or Gluconobacter can protect the worm from pathogen infection and killing in distinct ways5,6,11,13,14. A specific isolate of the genus Comamonas influences nematode dietary response, development, lifespan, and fertility15,16,17. Providencia bacteria produce the neuromodulator tyramine and thereby modulate host nervous system activity and resulting behavioral responses18. A set of different naturally associated bacteria were demonstrated to affect population growth rate, fertility, and behavioral responses5,6,9,11,19.
To date, the exact diversity and consistency of the native C. elegans microbiota across habitats and geographic locations are not fully understood, and further associations between the worm and microbes from its environment remain to be uncovered. Several previous studies used bacterial strains isolated from some soil environment, natural C. elegans habitats, or from mesocosm experiments (i.e., lab-based environments that recreate natural habitats) with C. elegans laboratory strains4,5,20. Even though these studies obtained new insights into the influence of microbes on specific nematode traits (e.g., nematode metabolism21), the relevance of these interactions for C. elegans biology in nature is unclear. Therefore, this manuscript describes the methods to directly isolate C. elegans from nature and to isolate and subsequently characterize the naturally associated microbes from both single worms and groups of worms. The described methods are an updated and improved version of the procedures used previously for the isolation and characterization of natural C. elegans and its native microbiota2,6,7. Considering that C. elegans is widely found in decomposing plant matter across the globe (especially in rotting fruits, temperate regions, and in autumn)1,2,22,23,24,25, this protocol can be applied by any lab whenever there is interest in relating C. elegans traits to naturally associated microbes and thus a more naturally relevant context. The latter is pivotal for a full understanding of the nematode's biology because it is known from a diversity of other host systems that the associated microbiota can affect diverse life history characteristics26, an aspect which is currently largely neglected in the multitude of C. elegans studies across almost all life science disciplines.
Protocol
1. Preparation of buffers and media
- Prepare S-buffer by adding 5.85 g of NaCl, 1.123 g of K2HPO4, 5.926 g of KH2PO4, and 1 L of deionized H2O to a flask and autoclave for 20 min at 121 °C.
- Prepare a viscous medium by adding S-buffer containing 1.2% (w/v) hydroxymethylcellulose (the substance causing viscosity of the medium), 5 mg/mL cholesterol, 1 mM MgSO4, 1 mM CaCl2, and 0.1% (v/v) acetone. Autoclave and stir the viscous medium until it is completely homogeneous.
NOTE: This can take multiple hours. Also, S-buffer can be directly prepared and added to the viscous medium without prior sterilization. - Prepare M9-buffer by adding 3 g of KH2PO4, 6 g of NA2HPO4·2 H2O, 5 g of NaCl, and 1 L of deionized H2O to a 1 L flask. Autoclave the solution, and after cooling, add 1 mL of 1 M MgSO4 (123.24 g of MgSO4·7H2O in 500 mL of deionized H2O, filter sterilized).
- Prepare 10% (v/v) Triton X-100 stock solution by mixing 5 mL of Triton X-100 with 45 mL of M9-buffer. Filter-sterilize the solution using a 0.2 µm filter.
- Prepare M9-buffer with Triton X-100 (M9-T) by adding 2.5 mL of the 10% (v/v) Triton X-100 stock solution to 1 L of M9-buffer after autoclaving to obtain 0.025% (v/v) M9-T.
- Prepare 30% (v/v) glycerol in S-buffer by mixing 15 mL of sterile 100% glycerol and 35 mL of sterile S-buffer in a 50 mL tube.
2. Preparation of environmental samples (Figure 1)
- Collect environmental samples like compost or rotten fruits and place each sample in an individual plastic bag, tube, or other clean container.
NOTE: In order to attract nematodes, apples can be placed on compost several weeks prior to the sampling. - Spread pieces of the environmental sample evenly in an empty, sterile, 9 cm Petri dish.
NOTE: Samples with higher levels of decay are more likely to contain C. elegans. Optionally, a Petri dish filled with peptone-free agar medium (PFM) can be used to increase contrast. - Cover the sample carefully with approximately 20 mL of sterile viscous medium.
NOTE: Nematodes float to the surface within 1-2 h. The viscous medium slows down the movement of the worms and makes it easier to sample them. Sterile M9-buffer can be used as an alternative, however, worm movement will be faster.
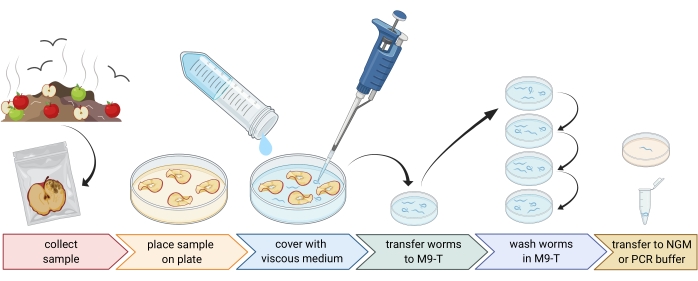
Figure 1: Isolation of nematodes from substrates. Substrate samples are placed in empty Petri dishes and covered with viscous medium to flush out nematodes. Nematodes are transferred to M9-T and repeatedly washed to remove bacteria from the outside. Individual nematodes can be used for DNA isolation, isolation of associated bacteria, or placed on agar plates to culture worm populations. Figure created with BioRender.com. Please click here to view a larger version of this figure.
3. Isolation of Caenorhabditis nematodes (Figure 1)
- Search for Caenorhabditis nematodes using a dissecting microscope, following the guidelines presented in the WormBook chapter on the isolation of C. elegans and related nematodes by Barrière and Félix27.
NOTE: Caenorhabditis hermaphrodites/females have a gut of light brown color (under transmitted illumination). Moreover, the gut cells have large cell nuclei, which are visible as white dots. By contrast, other nematode species often have a dark brown gut and may show anteroposterior asymmetry in pigment intensity and usually no visible nuclei. The vulva of adult Caenorhabditis hermaphrodites/females is found in the middle of the animal. This vulva localization is not always shown by other nematode taxa. Caenorhabditis nematodes possess two pharyngeal bulbs, which are both visible with sufficient magnification and transmitted illumination and contrast with many other nematode taxa. Caenorhabditis nematodes have a pointed tail, whereas some other nematodes have a round tail. - Collect the Caenorhabditis nematodes under a dissecting microscope in as little liquid as possible using a 20-100 µL pipette. Transfer the collected nematodes directly to 1-3 mL of sterile M9-T in a sterile 3 cm Petri dish for washing the worms to remove non-attached microbes (step 3.3) or, alternatively, transfer individual worms onto a plate with nematode growth medium (NGM) to establish a worm population (step 3.4).
- Wash the nematodes to remove microbes from the outside of the nematode.
NOTE: This protocol enriches for gut bacteria but does not completely eliminate bacteria sticking to the worm cuticle.- Incubate the nematodes for 10-15 min in M9-T.
- Pipette the nematodes in as little liquid as possible into 1-3 mL of fresh M9-T in a new sterile 3 cm Petri dish.
- Repeat the incubation and transfer the nematodes to fresh M9-T two more times.
NOTE: The following steps can be done either with individual worms or with worm populations. The worms can now be used for the characterization of C. elegans and associated microbes, as well as the isolation of bacteria (steps 4 and 5). Alternatively, they can be transferred individually to NGM plates to establish a worm population (step 3.4).
- To obtain a worm population, pipette an individual nematode to a NGM plate.
NOTE: Only hermaphroditic nematodes such as C. elegans or C. briggsae are able to produce offspring from single worms. However, single worms of other species may still produce offspring if they were already mated.- Naturally isolated nematodes usually contain bacteria in their gut, which they shed onto the NGM plates, where these bacteria grow and are available as food for C. elegans. Do not add separate food organisms like the standard laboratory food organism, E. coli strain OP50.
NOTE: Rearing worms on plates influences the composition of the associated bacterial community, yet the composition is still comparable to that of the natural C. elegans isolates6,7. - Allow the nematodes to proliferate for up to 10 days at the appropriate temperature (e.g., 15-20 °C for temperate locations). Freeze these nematodes (step 3.5) or use them for the characterization of C. elegans and associated microbes as well as the isolation of microbes (steps 4 and 5).
- Naturally isolated nematodes usually contain bacteria in their gut, which they shed onto the NGM plates, where these bacteria grow and are available as food for C. elegans. Do not add separate food organisms like the standard laboratory food organism, E. coli strain OP50.
- Freezing nematodes for long-term storage
- Leave the nematodes on plates until the food bacteria have disappeared, and there are mainly small larval stages on the plates. Wash the worms off the plates in 1.5 mL of S-buffer.
- Mix 500 µL of worm-containing S-buffer thoroughly with 500 µL of 30% (v/v) glycerol in S-buffer in a sterile 2 mL tube. Freeze the tubes promptly at -80 °C for long-term storage, otherwise the glycerol may harm the nematodes.
4. Preparation of the worms for molecular identification of C. elegans and microbes
- For unbiased identification of nematode-associated microbes, prepare a 96-well plate with three sterile 1 mm beads, 19.5 µL of PCR-buffer, and 0.5 µL of Proteinase K (20 mg/mL) per well. Pipette an individual, washed nematode to each well, transferring as little liquid as possible.
- Worm populations can also be used. For this, wash the worms from the plates with M9-buffer and transfer ~300 µL of worm-containing M9-buffer to 2 mL tubes with 10-15 beads.
- Break up the nematodes using a bead homogenizer (e.g., bead-beating for 3 min at 30 Hz). Centrifuge the plate or tubes briefly to get the liquid to the bottom (e.g., for 10 s at 8000 x g at room temperature [RT]).
- Identification of C. elegans
- Isolate the DNA of individual nematodes by heating the samples in a PCR cycler for 1 h at 50 °C and 15 min at 95 °C. Isolate the DNA of worm populations using any isolation method of choice (example protocols of different isolation methods using commercial kits7,9). Freeze the DNA at -20 °C for long-term storage.
- For the identification of C. elegans, use the DNA and the primer pair nlp30-F (Table 1, 5'-ACACATACAACTGATCACTCA-3') and nlp30-R (Table 1, 5'-TACTTTCCCCATCCGTATC-3') in a PCR, following the instructions of a Taq supplier of choice.
- Use the following PCR conditions: initial denaturation step at 95 °C for 2 min, followed by 35 cycles of 95 °C for 45 s, 55 °C for 30 s, 72 °C for 1 min, and a final elongation step at 72 °C for 5 min. C. elegans produces a 154 bp PCR product.
- Characterize the nematode-associated bacteria via 16S amplicon sequencing of the V3-V4 region, using the isolated DNA.
- Prepare a 16S library with the 16S primers of choice and follow the library preparation kit protocol. One option is to use the primers 341F (Table 1, 5'-CCTACGGGNGGCWGCAG-3') and 806R (Table 1, 5'-GACTACHVGGGTATCTAATCC-3') covering the V3-V4 region of the 16S rRNA gene, which results in sequences that can be classified with standard databases with good resolution7.
NOTE: The amount of input DNA is critical in this step. DNA acquired from single worms is going to be much less than that obtained from worm populations. For single worms, it might be necessary to increase the amount of input DNA or to increase the amount of PCR cycles7. - Libraries can be sequenced on a sequencing platform using a suitable sequencing kit.
NOTE: Here, the MiSeq platform is used with a suitable MiSeq Reagent Kit. The reagents are improved constantly and should be chosen to the newest standards.
- Prepare a 16S library with the 16S primers of choice and follow the library preparation kit protocol. One option is to use the primers 341F (Table 1, 5'-CCTACGGGNGGCWGCAG-3') and 806R (Table 1, 5'-GACTACHVGGGTATCTAATCC-3') covering the V3-V4 region of the 16S rRNA gene, which results in sequences that can be classified with standard databases with good resolution7.
5. Isolation and cultivation of nematode-associated bacteria (Figure 2)
- To isolate bacteria, prepare a 96-well plate with three sterile 1 mm beads, 20 µL of M9-buffer per well, and pipette an individual, washed nematode to each well, transferring as little liquid as possible.
NOTE: Alternatively, worm populations can be washed from the plates with M9-buffer and ~300 µL of worm-containing M9-buffer can be transferred to 2 mL tubes with 10-15 beads. In all cases, the worms should come from a culture that was identified to be C. elegans using steps 4.1-4.4. - Break up the nematodes using a bead homogenizer (e.g., bead-beating for 3 min at 30 Hz). Centrifuge the plate or tubes briefly to get the liquid to the bottom (e.g., for 10 s at 8000 x g at RT).
NOTE: The usage of this method led to the isolation of bacterial taxa similar to those revealed by 16S rDNA amplicon sequencing7, suggesting that the described bead-beating leaves most bacterial cells intact. - Collect the supernatant, serially dilute it at 1:10, and plate up to 100 µL onto 9 cm agar plates.
- To ensure that most bacteria can be cultivated, use a variety of alternative media with different nutrient compositions, including diluted trypticase soy agar (TSA, 1:10 dilution), MacConkey agar, Sabouraud glucose agar, potato dextrose agar, or yeast peptone dextrose agar.
- Incubate the plates at the average temperature conditions of the sampling location (e.g., temperatures between 15-20 °C for temperate locations) for 24-48 h.
- Use the standard three-streak technique and a sterile loop to obtain pure bacterial cultures (Figure 2).
- Pick a single colony from a plate using a sterile loop or toothpick and streak it out onto a new agar plate containing the same agar medium as used for purification. Make sure to use only roughly 1/3 of the plate.
- Either use a new sterile loop or sterilize a reusable loop and drag it through the first streak to create a second streak on another 1/3 of the same plate.
- Repeat this step by dragging a sterile loop through the second streak.
- Incubate the plate at the same growth conditions used for isolation. This technique must result in single colonies growing in the area of the third streak.
NOTE: It might be necessary to repeat the purification step multiple times as natural isolates tend to form biofilms and/or aggregates.
- Grow the pure colonies in a liquid medium (of the same type as the agar medium) using the same temperature and growth time as above (step 5.4)
- Prepare bacteria stocks by adding 300 µL of the bacterial culture to 200 µL of 86% (v/v) glycerol (in the respective growth medium, e.g., TSB) and pipette up and down to mix properly. Alternatively, prepare DMSO stocks by mixing 50 µL of bacterial culture with 50 µL of 7% (v/v) DMSO. Freeze at -80 °C for long-term storage.
- Characterizing bacteria using sequencing of the full 16S ribosomal RNA gene
- Extract the bacterial DNA from pure liquid cultures using a suitable technique (e.g., a DNA extraction kit; from experience, a CTAB-based extraction protocol works very well22).
- Amplify the 16S rRNA gene using the primers 27F (Table 1, 5'-GAGAGTTTGATCCTGGCTCAG-3') and 1495R (Table 1, 5'-CTACGGCTACCTTGTTACGA -3')28 and the following PCR conditions: 95 °C, 2 min, 22x (95 °C, 30 s; 55 °C, 30 s; 72 °C, 100 s), and a final extension period at 72 °C, 5 min.
- In order to acquire the full sequences, additionally use two internal sequencing primers, such as 701F (Table 1, 5'-GTGTAGCGGTGAAATGCG-3') and 785R (Table 1, 5'-GGATTAGATACCCTGGTAGTCC-3')6.
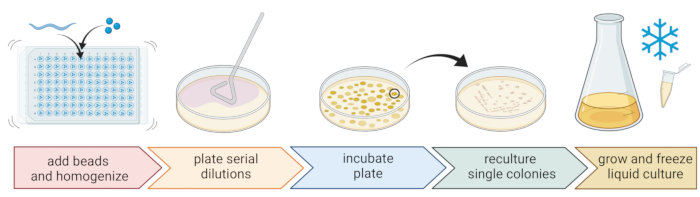
Figure 2: Species identification and isolation of individual bacteria. Individual nematodes are broken up using a bead homogenizer, and DNA is isolated for species determination via PCR or sequencing. Alternatively, the broken-up nematode material is serially diluted and plated onto growth medium plates. Plates are incubated until bacterial colonies appear, and single colonies are streaked to new plates to obtain pure cultures. Single colonies of the pure cultures are used to grow liquid bacterial cultures for the preparation of bacterial stocks for long-term storage at -80 °C. Figure created with BioRender.com. Please click here to view a larger version of this figure.
Results
The nematode C. elegans is frequently found in decomposing fruits, such as apples, and also compost samples. In Northern Germany, C. elegans as well as congeneric species (particularly C. remanei but also C. briggsae) are mainly found from September until November2. The nematodes are most commonly found in decomposing plant matter, especially rotting fruits such as apples or pears, and also compost, particularly material that shows a high grade of decomposition....
Discussion
The nematode Caenorhabditis elegans is one of the most intensively studied model organisms in biological research. It was introduced by Sydney Brenner in the 1960s, originally for understanding the development and the function of the nervous system29. Since then, C. elegans has become a powerful model for studying fundamental processes across all biological disciplines, including behavioral biology, neurobiology, aging, evolutionary biology, cell biology, developmental biology, a...
Disclosures
We declare that we have no conflict of interest.
Acknowledgements
We acknowledge financial support from the German Science Foundation (projects A1.1 and A1.2 of the Collaborative Research Center 1182 on the Origin and Function of Metaorganisms). We thank the members of the Schulenburg lab for their advice and support.
Materials
| Name | Company | Catalog Number | Comments |
| COMSOL | COMSOL | multiphysics simulation software |
References
- Schulenburg, H., Félix, M. -. A. The natural biotic environment of Caenorhabditis elegans. Genetics. 206 (1), 55-86 (2017).
- Petersen, C., Dirksen, P., Prahl, S., Strathmann, E. A., Schulenburg, H. The prevalence of Caenorhabditis elegans across 1.5 years in selected North German locations: the importance of substrate type, abiotic parameters, and Caenorhabditis competitors. BMC Ecology. 14 (1), 4 (2014).
- Petersen, C., et al. Travelling at a slug's pace: possible invertebrate vectors of Caenorhabditis nematodes. BMC Ecology. 15 (1), 19 (2015).
- Berg, M., et al. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. The ISME Journal. 10 (8), 1998-2009 (2016).
- Samuel, B. S., Rowedder, H., Braendle, C., Félix, M. -. A., Ruvkun, G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proceedings of the National Academy of Sciences. 113 (27), 3941-3949 (2016).
- Dirksen, P., et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biology. 14 (1), 38 (2016).
- Johnke, J., Dirksen, P., Schulenburg, H. Community assembly of the native C. elegans microbiome is influenced by time, substrate and individual bacterial taxa. Environmental Microbiology. 22 (4), 1265-1279 (2020).
- Zhang, F., et al. Caenorhabditis elegans as a model for microbiome research. Frontiers in Microbiology. 8, 485 (2017).
- Dirksen, P., et al. CeMbio - The Caenorhabditis elegans microbiome resource. G3 Genes|Genomes|Genetics. 10 (9), 3025-3039 (2020).
- Zimmermann, J., et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. The ISME Journal. 14 (1), 26-38 (2019).
- Kissoyan, K. A. B., et al. Exploring effects of C. elegans protective natural microbiota on host physiology. Frontiers in Cellular and Infection Microbiology. 12, 775728 (2022).
- Zhang, F., et al. Natural genetic variation drives microbiome selection in the Caenorhabditis elegans gut. Current Biology. 31 (12), 2603-2618 (2021).
- Berg, M., et al. TGFβ/BMP immune signaling affects abundance and function of C. elegans gut commensals. Nature Communications. 10 (1), 604 (2019).
- Kissoyan, K. A. B., et al. Natural C. elegans microbiota protects against infection via production of a cyclic lipopeptide of the viscosin group. Current Biology. 29 (6), 1030-1037 (2019).
- Watson, E., MacNeil, L. T., Arda, H. E., Zhu, L. J., Walhout, A. J. M. Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell. 153 (1), 253-266 (2013).
- Watson, E., et al. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell. 156 (4), 759-770 (2014).
- MacNeil, L. T., Watson, E., Arda, H. E., Zhu, L. J., Walhout, A. J. M. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 153 (1), 240-252 (2013).
- O'Donnell, M. P., Fox, B. W., Chao, P. -. H., Schroeder, F. C., Sengupta, P. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature. 583 (7816), 415-420 (2020).
- Snoek, B. L., et al. A multi-parent recombinant inbred line population of C. elegans allows identification of novel QTLs for complex life history traits. BMC Biology. 17 (1), 24 (2019).
- Avery, L., Shtonda, B. B. Food transport in the C. elegans pharynx. Journal of Experimental Biology. 206 (14), 2441-2457 (2003).
- Zhang, J., et al. A delicate balance between bacterial iron and reactive oxygen species supports optimal C. elegans development. Cell Host & Microbe. 26 (3), 400-411 (2019).
- Petersen, C., et al. Ten years of life in compost: temporal and spatial variation of North German Caenorhabditis elegans populations. Ecology and Evolution. 5 (16), 3250-3263 (2015).
- Félix, M. -. A., Duveau, F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biology. 10 (1), 59 (2012).
- Barrière, A., Félix, M. -. A. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics. 176 (2), 999-1011 (2007).
- Dolgin, E. S., Félix, M. -. A., Cutter, A. D. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity. 100 (3), 304-315 (2008).
- Douglas, A. E. Simple animal models for microbiome research. Nature Reviews Microbiology. 17 (12), 764-775 (2019).
- Barrière, A., Félix, M. -. A. Isolation of C. elegans and related nematodes. WormBook. , 1-19 (2014).
- Weisburg, W. G., Barns, S. M., Pelletier, D. A., Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 173 (2), 697-703 (1991).
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics. 77 (1), 71-94 (1974).
- Crombie, T. A., et al. Local adaptation and spatiotemporal patterns of genetic diversity revealed by repeated sampling of Caenorhabditis elegans across the Hawaiian Islands. Molecular Ecology. 31 (8), 2327-2347 (2022).
- Haber, M. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: Evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Molecular Biology and Evolution. 22 (1), 160-173 (2004).
- Watson, E., et al. Metabolic network rewiring of propionate flux compensates vitamin B12 deficiency in C. elegans. eLife. 5, 17670 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved