Method Article
Ultrastructural Localization of Endogenous LC3 by On-Section Correlative Light-Electron Microscopy
In This Article
Summary
Here, we present a protocol for optimized on-section correlative light-electron microscopy based on endogenous, fluorescent labeling as a tool to investigate the localization of rare proteins in relation to cellular ultrastructure. The power of this approach is demonstrated by ultrastructural localization of endogenous LC3 in starved cells without Bafilomycin treatment.
Abstract
The visualization of autophagic organelles at the ultrastructural level by electron microscopy (EM) is essential to establish their identity and reveal details that are important for understanding the autophagic process. However, EM methods often lack molecular information, obstructing the correlation of ultrastructural information obtained by EM to fluorescence microscopy-based localization of specific autophagy proteins. Furthermore, the rarity of autophagosomes in unaltered cellular conditions hampers investigation by EM, which requires high magnification, and hence provides a limited field of view.
In answer to both challenges, an on-section correlative light-electron microscopy (CLEM) method based on fluorescent labeling was applied to correlate a common autophagosomal marker, LC3, to EM ultrastructure. The method was used to rapidly screen cells in fluorescence microscopy for LC3 labeling in combination with other relevant markers. Subsequently, the underlying ultrastructural features of selected LC3-labeled spots were identified by CLEM. The method was applied to starved cells without adding inhibitors of lysosomal acidification.
In these conditions, LC3 was found predominantly on autophagosomes and rarely in autolysosomes, in which LC3 is rapidly degraded. These data show both the feasibility and sensitivity of this approach, demonstrating that CLEM can be used to provide ultrastructural insights on LC3-mediated autophagy in native conditions-without drug treatments or genetic alterations. Overall, this method presents a valuable tool for ultrastructural localization studies of autophagy proteins and other scarce antigens by bridging light microscopy to EM data.
Introduction
Autophagy is a key process for the clearance and recycling of cytoplasmic proteins and organelles. The process of macro-autophagy (hereafter called autophagy) involves the formation of double-membrane organelles, autophagosomes, which allows cells to enclose cytoplasmic molecules and organelles for lysosomal degradation. Autophagy occurs at a basal level in most cells and is upregulated in response to cellular conditions, such as starvation or cellular stress. Autophagy either occurs in a substrate-specific manner, targeting specific structures or proteins for degradation, or as a non-selective bulk process encompassing parts of the cytosol. In selective autophagy, autophagosomes are formed by the conjugation of Atg8-family proteins (microtubule associated proteins 1A/B light chain 3A/B/C [LC3] and GABARAPs) to membranes derived from recycling endosomes, the Golgi, and/or endoplasmic reticulum (ER)1. LC3 recognizes autophagic cargo in the cytosol directly or via selective autophagy adaptors such as P62/SQSTM. New autophagic membranes can then be conjugated to LC3, expand, and fuse to form a completed double membrane enclosing the cargo-called the autophagosome. The autophagosome matures and eventually fuses with an endosome or lysosome, whereafter the autophagic cargo and adaptors are degraded2.
Studies on autophagosome formation, maturation, and fusion often make use of light microscopy technologies. Fluorescence microscopy of LC3 is generally used to assess the number and cellular localization of autophagosomes under different conditions. Furthermore, by coupling LC3 to pH-sensitive GFP and pH-stable RFP in a so-called tandem probe, the overall flux of autophagy can be measured in live cells as a function of GFP fluorescence loss3. These approaches are valuable tools for researchers to understand the role and mechanism of autophagy under different conditions. Another invaluable tool is electron microscopy (EM), which reveals the ultrastructure of autophagic organelles at different stages of autophagy4,5,6,7,8. To date, EM is still the method of choice to identify the precise stages of autophagosome formation by discriminating different autophagic membranes by morphology: phagophore (double membrane not fully closed), autophagosome (closed double membrane around cytosolic cargo), and autolysosome ([partial] loss of the inner autophagic membrane). Morphology without molecular information, however, can be prone to misidentification or ambiguity. Immuno-EM is the most comprehensive method for simultaneous molecular characterization and morphological classification of autophagic organelles. For example, immunogold labeling of LC3 on thawed cryosections allows the ultrastructural localization of LC3 and precise identification of LC3-marked organelles9.
A drawback of EM is the small field of view that comes with the high magnification required to observe the fine ultrastructure of autophagic membranes and, in the case of immuno-EM, to locate the label that marks the protein of interest. Due to their scarcity and low protein levels, this generally hampers the quantitative EM analysis of autophagosomes. To increase the number of autophagosomes, cells are often starved and treated with Bafilomycin A1 (BafA1), an inhibitor of lysosomal acidification and degradation. Without BafA1 treatment, the search for autophagosomes by EM is time-intensive, due to the scarcity of these organelles. The method presented in this manuscript addresses this issue through fluorescent labeling and imaging of endogenous LC3 on thawed cryosections in a fluorescence microscope before further preparation for EM. The fluorescent images then guide the search for LC3-labeled structures in the EM. After collection, EM images are correlated with the fluorescence images to add molecular information—the presence of LC3—to the ultrastructure of the cell. This 'on-section CLEM' method greatly increases the ability to find LC3-labeled structures, especially in untreated conditions, for subsequent identification and classification by EM.
This method was applied to starved hepatoblastoma-derived HEPG210 cells to find autophagosomes in unaltered (i.e., no BafA1 was used) conditions. Relatively few fluorescent puncta (less than one per cell profile in a 90 nm section) were found, which is in agreement with the high turnover of LC311. This sparsity of LC3-puncta emphasized the value of CLEM; by selecting regions with several fluorescent puncta for imaging in the EM, LC3-positive organelles were found and characterized in a much more effective manner than through conventional immuno-EM. This revealed that the majority of LC3-positive organelles were autophagosomes, as defined by their morphology, which is in contrast to the results obtained in BafA1-treated cells, where autolysosomes are more common9. These data show that with on-section CLEM, autophagy can be studied at the ultrastructural level without the need to inhibit autophagic flow.
Protocol
1. Preparation of tools and reagents
NOTE: For required reagents, buffers, and solutions, see Supplemental File 1 or 12 for more information. For details related to all materials, reagents, equipment, and software used in this protocol, see the Table of Materials.
- Fixatives
- Prepare 0.2 M phosphate buffer (PB) or 0.2 M PIPES, HEPES, EGTA, MgSO4 (PHEM) buffer, as described in Supplemental File 1, to use as a base for fixative solutions.
NOTE: Fixatives are routinely buffered in 0.1 M PB or PHEM buffer to buffer against acidification caused by the aldehyde reaction with the biological material. - As the quality of the paraformaldehyde (PFA) is key to reliable fixation of the samples' ultrastructure, use EM-grade PFA. To follow this protocol, use 16% stock solutions, prepared from high-quality PFA prills (see Supplemental File 1).
CAUTION: Paraformaldehyde is a hazardous chemical (hazard statements H228, H301, H302, H311, H314, H315, H317, H318, H331, H332, H335, H341, H350). When working with PFA, wear protective equipment (gloves, lab coat, and protective glasses) and work in a chemical hood. Waste that contains PFA should be collected and disposed of according to the institutes' guidelines and regulations. - Combine 10 mL of 0.2 M PB, 5 mL of 16% PFA (in demineralized water [dH2O]), and 5 mL of dH2O to prepare a fixative solution of 4% PFA.
- Optional: Adding 0.02%-0.5% glutaraldehyde (GA) to the fixative solution from step 1.1.3 improves the preservation of the ultrastructure, but reduces the antigenicity of the sample toward many antibodies.
NOTE: When GA fixation is desired, use EM-grade GA from a suitable supplier.
CAUTION: Glutaraldehyde is a hazardous chemical (hazard statements H301, H302, H314, H317, H330, H332, H334, H335, H400, H411). Work in a chemical hood and wear protective equipment (gloves, lab coat, and protective glasses) when manipulating GA. Waste containing GA should be collected and disposed according to the institutes' guidelines and regulations.
- Prepare 0.2 M phosphate buffer (PB) or 0.2 M PIPES, HEPES, EGTA, MgSO4 (PHEM) buffer, as described in Supplemental File 1, to use as a base for fixative solutions.
- Tools and materials
- Scratch the surface of aluminum sample holder pins and sonicate them in ethanol 3 x 10 min to remove metal remnants and ensure optimal adherence when gelatin-embedded cell blocks are mounted on the pins.
- Use a storage canister suitable to store the aluminum sample holder pins with their samples in liquid nitrogen (LN2).
- Make a manipulator by affixing a single hair or eyelash to the end of a wooden skewer using nail polish.
- Make a pickup loop. Bend a 0.3 mm thick stainless-steel wire around a 3 mm diameter round bar and twist the ends together, forming a loop on one end. Insert the twisted ends into a pipette tip. Insert a wooden skewer from the other end and affix with glue or resin.
NOTE: A pickup loop is also commercially available (see Table of Materials). - To prepare the grid-drying loops, follow the same steps for making pickup loops: form a stainless-steel wire in a 4 mm loop and affix onto a large pipette tip with glue or resin.
- Coat the grids using a thin supportive film such as formvar (protocol in Supplemental File 1). Before use, coat the grids with a thin layer of carbon.
NOTE: Ready-to-use grids are commercially available (see Table of Materials). Formvar-coated grids can be stored indefinitely at room temperature (RT); carbon-coated grids can be stored for several months at RT. - Prepare clean glass slides and large coverslips (24 mm x 24 mm is ideal with 25 mm wide glass slides), as in 13.
2. Fixation and sample preparation
- Fixation
- Use the fixative prepared in step 1.1.3 (4% PFA in 0.1 M PB). For adherent cell lines, culture 1-5 × 106 cells in 6 cm dishes. Add the fixative to the culture medium at a 1:1 ratio and incubate the sample for 5 min at RT. Then, replace the medium-fixative mixture with fixative only and incubate for 2 h at RT.
NOTE: Exact cell counts, confluence, and culture conditions may vary according to the model system used. - Store the samples overnight or for up to 3-4 weeks in 0.5% PFA in 0.1 M PB at 4 °C.
NOTE: GA can be added to the fixation (see step 1.1.4) and the fixation length can be changed to find an optimal balance between the preservation of morphology and antigenicity, which differs per specimen and labeling. For more information, see14.
- Use the fixative prepared in step 1.1.3 (4% PFA in 0.1 M PB). For adherent cell lines, culture 1-5 × 106 cells in 6 cm dishes. Add the fixative to the culture medium at a 1:1 ratio and incubate the sample for 5 min at RT. Then, replace the medium-fixative mixture with fixative only and incubate for 2 h at RT.
- Sample embedding
- Wash the dish with fixed cells 3x with PBS at RT. Then, replace with PBS containing 0.15% glycine and incubate for 10 min at RT.
- Replace the PBS containing 0.15% glycine with 1% gelatin in PBS prewarmed to 37 °C, and scrape and transfer the cells in 1% gelatin to a microcentrifuge tube. Pellet the cells at 6,000 × g for 1 min at RT in a microcentrifuge. Then, remove the 1% gelatin without disturbing the pellet and add 12% gelatin warmed to 37 °C. Resuspend the cell pellet by gently pipetting up and down with pipette tips or glass Pasteur pipettes prewarmed to 37 °C.
- Incubate at 37 °C for 10 min; then, pellet the cells at 6,000 × g for 1 min. Solidify the gelatin on ice for 30 min.
- To remove the gelatin-embedded cells from the tube, cut the tube end containing the pellet off from the rest of the tube with a razor blade. Then, perpendicular to the first cut, cut the tube end with the cell pellet in half.
- Incubate the two tube-end halves containing the gelatin-embedded cell pellet in 2.3 M sucrose for 10 min at 4 °C. This causes the gelatin-embedded cell pellet halves to shrink slightly and dislodge from the plastic tube.
NOTE: The gelatin-embedded cell pellets should be kept at 4 °C or ice-cold as much as possible to avoid the 2.3 M sucrose from becoming too viscous and the gelatin too soft. During manipulation of the gelatin-embedded cell pellets in the next steps, work with only one sample at a time and keep the others on ice, or work in a cold (~4 °C) room. Avoid overheating of the gelatin-embedded cell pellets by sunlight, hot microscope lamps, or other sources of heat. - Remove the tube halves with the gelatin-embedded cell pellet from the 2.3 M sucrose. Then, remove the gelatin-embedded cell pellet halves from the plastic tube halves with tweezers. Manually cut the pellet to blocks of an appropriate size (~1 mm3) with a razor blade. Use a stereo dissecting microscope to magnify the subject during the cutting.
- Infuse the gelatin-embedded cell blocks with 2.3 M sucrose for 3-16 h, turning end-over-end in a rotor at 4 °C.
- Place a gelatin-embedded cell block on an aluminum sample holder pin (see step 1.2.1). Leave enough 2.3 M sucrose around the edges of the block so that it forms a thin 'collar' between the block and the pin. Avoid too much 2.3 M sucrose covering the top of the block. Snap-freeze and store in LN2.
3. Sectioning
- Trimming (see also12)
- Take a pin with a block of gelatin-embedded cells out of LN2 storage and place it inside a cryomicrotome set at -80 °C.
- Trim the front of the block to flatten its surface and obtain ~250 nm sections. Dip the 3 mm loop in pickup solution (1:1 2.3 M sucrose and 2% methylcellulose), insert the loop into the cryochamber of the microtome, and wait until ice starts to form in the droplet (typically 5-7 s). Then, immediately pick up the section by quickly but gently pressing the droplet against them. Remove the loop from the cryochamber, wait until the droplet has thawed completely, and press the droplet on a glass slide.
- Check the cell orientation by toluidine blue staining of the sections.
- Place a drop of toluidine blue solution (see Supplemental File 1) on top of the sections on a glass slide and dry on an 80 °C heating plate until the edges of the drop are dry.
- Remove the glass slide from the heating plate and gently rinse the toluidine blue away with dH2O, collecting it in a suitable waste container.
- Dry the glass slide and check the cell orientation in the sections through a simple, benchtop light microscope.
- Trim the sides of the block by sectioning 50-100 µm into the side of the sample block face with the knife corner. Trim four sides of the sample block face by rotating the sample holder 90° after trimming each side to create a protruding ~250 µm x 375 µm rectangle. Select the protruding area based on the cell orientation determined in the previous step.
- Sectioning and pickup
- Cool the cryomicrotome to -100 °C. Section a ribbon from the protruding rectangle, the sections being 70-90 nm thick and with a silvery-golden sheen. Guide the sections away from the diamond knife's edge with a hair on a stick (see section 1.2.3) to create a long (2-5 mm) ribbon.
- Once a suitable ribbon has formed, stop sectioning to pick up the ribbon. Dip the 3 mm pickup loop in 2.3 M sucrose and 2% methylcellulose mixed 1:1, insert the loop into the cryochamber of the microtome, and wait until the droplet starts to freeze (typically 5-7 s). Then, immediately pick up the sections by quickly but gently pressing the droplet against them. Remove the loop from the cryochamber, wait until the droplet has thawed completely, and press the droplet on a prepared grid (step 1.2.6).
NOTE: Grids with sections can be stored at 4 °C for several months.
4. Labeling and light microscopy
- Labeling
- Place the grids with sections (Figure 1A) section-side down on ~1 mL of PBS in a small dish or multi-well plate. Incubate at 37 °C for 30 min.
NOTE: This step removes the gelatin that is in between the cells; gelatin is not needed after sectioning and interferes with the remaining protocol. - Process the grids section-side down on ~75 µL droplets on parafilm (see Figure 1B). Start with PBS + 0.15% glycine washes (3 x 2 min) at RT. Then, incubate the grids with 0.1% bovine serum albumin (BSA)-c + 0.5% fish skin gelatin (FSG) in PBS for 10 min at RT as the blocking step. Dilute primary antibodies in 0.1% BSA-c + 0.5% FSG in PBS and incubate the grids on ~10 µL droplets of this solution for 1 h at RT (Figure 1C).
- Wash the grids in 0.1% BSA in PBS 5x at RT. Then, dilute secondary antibodies and 4',6-diamidino-2-phenylindole (DAPI; 10 µg/mL) in 0.1% BSA-c + 0.5% FSG in PBS and incubate the grids on ~10 µL droplets of this solution for 30+ min at RT (Figure 1C). Wash the grids in PBS 5x at RT.
NOTE: Optionally, a secondary antibody can be labeled with 5, 10, 15, or 20 nm colloidal gold particles conjugated to protein A (PAG) for localization of the protein of interest in EM. If this is desired, incubate the grids with PAG for 20 min at RT after step 4.1.3. Then, wash 5x with PBS at RT. Avoid the simultaneous use of multiple primary antibodies and take note of the reactivity of PAG to different species' IgGs to prevent unwanted cross-reactions. For more information, see12.
- Place the grids with sections (Figure 1A) section-side down on ~1 mL of PBS in a small dish or multi-well plate. Incubate at 37 °C for 30 min.
- Mounting samples for light microscopy
- Submerge the grids in 50% glycerol in dH2O 2 x 5 min at RT. Sandwich the grids between a glass slide and coverslip in 50% glycerol, one grid per coverslip, with sections facing the coverslip (Figure 1D).
NOTE: The quality of the labeling can deteriorate when grids are kept mounted in 50% glycerol for more than 30 min. It is therefore recommended to mount and image two or three grids at a time and leave the others on the secondary labeling solution.
- Submerge the grids in 50% glycerol in dH2O 2 x 5 min at RT. Sandwich the grids between a glass slide and coverslip in 50% glycerol, one grid per coverslip, with sections facing the coverslip (Figure 1D).
- Light microscopy
- Take a glass slide with sandwiched grids to a widefield microscope with an automated stage. Select a high-magnification (63x or 100x) oil objective. Create an image tileset of (part of) the ribbon of sections (Figure 1E).
NOTE: Some secondary antibodies can form fluorescent aggregates on grids, especially around folds or tears in the sections. Additionally, some cell types and tissues contain autofluorescent structures. If such issues are expected, it is advised to include a negative control grid not incubated with primary antibody.
- Take a glass slide with sandwiched grids to a widefield microscope with an automated stage. Select a high-magnification (63x or 100x) oil objective. Create an image tileset of (part of) the ribbon of sections (Figure 1E).
- Unmounting and EM contrasting
- Add 10 µL of dH2O to the side of the glass slide-coverslip sandwich and wait for capillary action to fill the glass-coverslip sandwich interface. Carefully remove the coverslip without mixing immersion oil into the glycerol. Retrieve the grids with tweezers and submerge in dH2O 3x at RT to wash off the 50% glycerol.
NOTE: Oil can interfere with uranyl staining and deteriorate EM contrast. - Carefully dry the back of the grid with lint-free tissue paper.
NOTE: If the sample was also labeled with colloidal gold particles, perform the following steps: place the grid with sections section-side down on PBS droplets and wash 2x at RT. Postfix in 1% GA for 5 min at RT (see caution note under 1.1.4). Wash in PBS 2x at RT. - Place the grids section-side down on dH2O droplets and wash 8x at RT.
- To stain the sections for contrast in EM, incubate with uranyl acetate (UA), pH 7, for 5 min at RT (Figure 1F).
- Before placing the grids, cool the UA:methylcellulose, pH 4, by placing droplets on parafilm on a metal plate on ice. Then, wash the grids with ice-cold UA:methylcellulose, pH 4, 2x and incubate with ice-cold UA:methylcellulose, pH 4, for 10 min (Figure 1F).
CAUTION: Uranyl acetate is a hazardous chemical (hazard statements H300, H330, H373, H411). In steps that require UA, work in a chemical hood and wear protective equipment (lab coat, gloves, and protective glasses). Collect and dispose of waste containing UA according to the institutes' guidelines and regulations. - Loop out the grids by inserting a grid-drying loop into the UA:methylcellulose droplet below the grid and gently lifting it until the grid is pulled off the droplet12. Blot the excess UA:methylcellulose away by touching the loop at a ~60° angle (sections facing down) onto lint-free filter paper (see Table of Materials) and dragging it slowly along the paper until no more UA:methylcellulose is absorbed. Then, place the loop with the grid in a suitable rack and let dry for >10 min at RT (Figure 1G).
- Add 10 µL of dH2O to the side of the glass slide-coverslip sandwich and wait for capillary action to fill the glass-coverslip sandwich interface. Carefully remove the coverslip without mixing immersion oil into the glycerol. Retrieve the grids with tweezers and submerge in dH2O 3x at RT to wash off the 50% glycerol.
5. EM
- Use the overview obtained by light microscopy to locate a region of interest (ROI) for imaging in the transmission electron microscope (TEM; Figure 1H). Annotate the ROI on the light microscopy dataset. Once a region is selected, obtain an image tileset at 20,000x-50,000x magnification in the TEM. Reconstruct the image tileset in postprocessing software15,16.
6. Correlation and analysis
- Load the light microscopy and EM dataset in suitable image processing software, such as ImageJ/Fiji17, the ec-CLEM plug-in in Icy18, or Photoshop. Crop and rotate the light microscopy dataset to match the EM tileset.
- Perform the correlation based on DAPI signal in fluorescence and nuclear outlines in EM (Figure 1I). Shift the images to overlay them precisely and perform the manual correlation accurately. To make the approach more exact, apply landmark-based correlation through, for example, the ec-CLEM plugin in Icy or the BigWarp plugin in ImageJ, to correlate the images through manual selection of corresponding points. A detailed, step-by-step protocol for correlation with ec-CLEM is available19.
NOTE: This approach also works well with the use of bimodal fiducial probes20,21.
- Perform the correlation based on DAPI signal in fluorescence and nuclear outlines in EM (Figure 1I). Shift the images to overlay them precisely and perform the manual correlation accurately. To make the approach more exact, apply landmark-based correlation through, for example, the ec-CLEM plugin in Icy or the BigWarp plugin in ImageJ, to correlate the images through manual selection of corresponding points. A detailed, step-by-step protocol for correlation with ec-CLEM is available19.
- Analyze the correlated images by selecting ROIs based on fluorescent signal in a suitable program (e.g., ImageJ). For quantitative analysis, create a collection of ROIs for all labeled organelles. Then, inspect the corresponding ultrastructure of the individual ROIs and classify them based on morphological elements.
Results
An optimized immuno-EM protocol for immuno-gold labeling of LC3 on ultrathin cryosections was recently published by De Maziere et al.9. This study included starved conditions without BafA1, in which LC3 was present but relatively rare and difficult to find by EM. An on-section CLEM method was introduced in a separate study, which uses the sensitivity of fluorescence labeling to visualize relatively rare and low expressed endogenous proteins and correlate this to EM ultrastructure14. Here, these two approaches are combined by the use of the optimized LC3 labeling protocol as part of a CLEM approach.
HEPG2 cells, liver-derived cells with relatively high levels of basal autophagy22, were starved in minimal medium (Earle's balanced salt solution [EBSS]) for 2 h prior to fixation in 4% PFA. This was followed by sample preparation by the Tokuyasu method of ultrathin cryosectioning (sections 1-3; see Slot and Geuze12), which is highly compatible with on-section CLEM14,23. Thawed cryosections were fluorescently labeled (protocol section 4 and Figure 1) using mouse anti-LC3 primary antibody9. Additionally, rabbit anti-LAMP1 was used to indicate endo-lysosomes, followed by anti-mouse AlexaFluor488 and anti-rabbit AlexaFluor568 secondary antibodies. Grids were sandwiched between a coverslip and glass slide and imaged at RT on a widefield microscope (100x 1.47 NA oil objective, sCMOS camera).
An advantage of fluorescence labeling of thin sections over conventional whole-cell IF is the increased resolution in Z, since the physical thickness of the section is 60-90 nm. With this improved Z resolution, the fluorescence labeling of LC3 and LAMP1 on thin sections reveals very little colocalization (Figure 2A). In cells treated with lysosomal inhibitors, such as BafA1, high colocalization occurs, since lysosomal-enclosed LC3 remains undegraded9. In untreated cells, LC3 is rapidly degraded upon contact with enzymatically active, LAMP1-positive lysosomes, and therefore co-localization is rare in these conditions. Generally, less than one LC3 punctum per cell profile was observed. This indicates that even in starved conditions, the turnover of autophagosomes is rapid, keeping autophagosome numbers low. It also highlights the importance of using CLEM to find the rare LC3-labeled structures, using the large field of view provided by light microscopy. Moreover, the higher sensitivity of fluorescence labeling in comparison to gold labeling enables the identification of more LC3-positive organelles than in conventional immuno-EM, further aiding their characterization.
After acquiring a full tileset of the ribbon of sections, the grids were retrieved from the microscope and post-stained for EM using UA and the loop-out method (protocol steps 4.4-4.6; Figure 1F,G). This 'loop-out' method ensures that a thin layer of UA:methylcellulose remains on the grid, which creates the desired contrast in the EM. The thickness of the layer depends on the speed and angle with which the UA:methylcellulose is blotted off onto the filter paper. Dragging the loop too quickly can leave too much UA:methylcellulose on the grid and darken the appearance of the sections in EM. Dragging too slowly can draw too much UA:methylcellulose away, resulting in too little staining and poor morphology, and risks the grid falling out of the loop. 'Oil slick' coloring (Figure 1G) on dry grids indicates a suitable UA:methylcellulose layer thickness.
After loop-out and drying, the grids were imaged in a TEM at ROIs selected by fluorescence. The IF and EM datasets were correlated by overlaying the DAPI signal to the outlines of nuclei visible in the EM, generating an integrated image containing information of both modalities.
Finding the same area in EM as selected in IF can be challenging. It is therefore recommended to keep an overview image of the IF tileset at hand while searching in the EM. Users should look for recognizable features in both modalities, such as folds or tears in the sections, grid bars, or arrangement of nuclei. It is also important to keep in mind that the sample can appear rotated and mirrored in EM. 'Finder grids' with specific features to identify areas can be used to ease correlation (see Table of Materials).
Correlation of the LC3-positive organelles to EM ultrastructure revealed that the different puncta represented distinct stages of autophagy (Figure 2B). Although the preservation of autophagosomal ultrastructure is challenging in cryosections, organelles with cytoplasmic content and double membranes were frequently observed (Figure 2C, arrows in organelles 1-5; Supplemental Figure S1), which are defining morphological features of autophagosomes. Interestingly, rather weak fluorescent spots were identified by EM as LC3-positive autolysosomes (Figure 2C, organelle 6; autophagic content is marked *), characterized by dense content and intraluminal vesicles. This showed that very small amounts of LC3 are visible in IF of ultrathin cryosections, and indicated that despite the degradative milieu, some LC3 is detectable in steady-state autolysosomes. However, the majority of LC3-positive puncta represented autophagosomes, whereas autolysosomes were very rare. This is in contrast with BafA1-treated cells, which primarily accumulate autolysosomes and not autophagosomes9.
In summary, this protocol describes an on-section CLEM method for linking molecular information obtained by fluorescence microscopy to the ultrastructure of EM. This method increases the sensitivity of immuno-EM, since only fluorophores are used for labeling and these generally yield more signal than EM probes. The method is especially suited for using ultrathin cryosections, in which high levels of specific fluorescence over negligible background staining can be obtained. By using fluorescence to screen for rare structures or events and correlating selected ROIs to EM, the EM operation time and associated costs can be greatly decreased. The sensitivity and feasibility of the method is demonstrated by the visualization of LC3 in untreated, starved cells, showing that LC3 predominantly associates to autophagosomes in these conditions, with very low levels visible in autolysosomes.
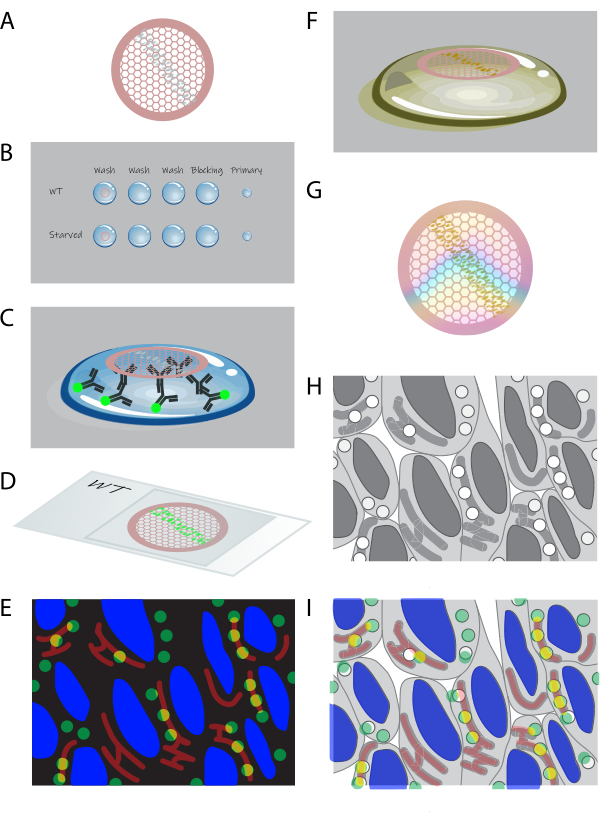
Figure 1: Schematic overview of on-section CLEM. (A) Cryosections from gelatin-embedded cells are collected on a formvar-coated copper grid. (B) Grids are processed section-down on droplets of the appropriate solutions. (C) Grids are labeled with primary and fluorescent secondary antibodies. (D) Grids are sandwiched between a coverslip and glass slide in 50% glycerol. (E) Fluorescence images are collected in a widefield microscope. (F) Grids are retrieved from the glass slide and further processed by uranyl staining for EM. (G) After drying, the grids can be imaged by TEM. (H) High magnification TEM image tileset is acquired from an area selected from fluorescence data. (I) Images from fluorescence microscopy and EM are correlated and overlaid. Please click here to view a larger version of this figure.
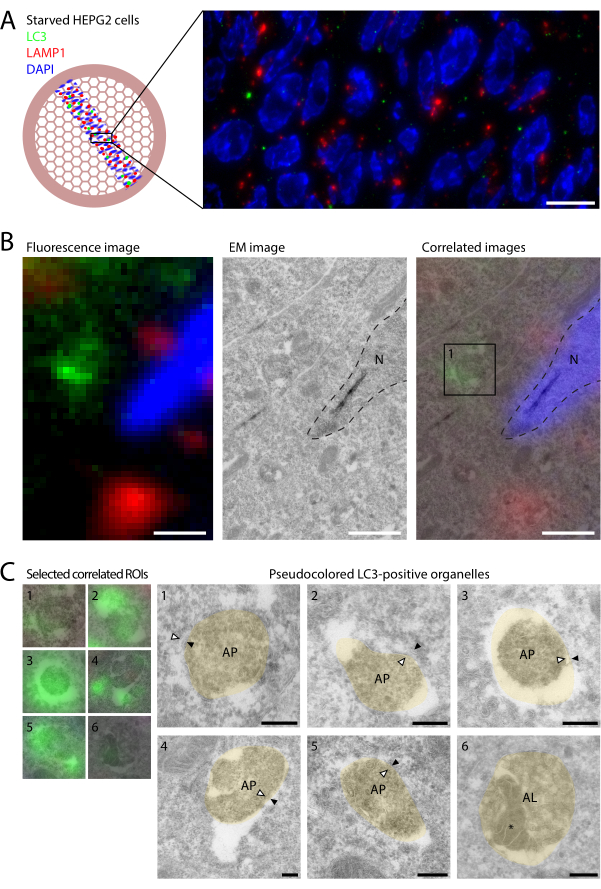
Figure 2: CLEM of LC3 and LAMP1 in starved HEPG2 cells. HEPG2 cells were starved for 2 h in EBSS prior to fixation with 4% PFA for 2 h. (A) IF imaging of LC3 (green) and LAMP1 (red) on sections reveals relatively few LC3 puncta and little colocalization with LAMP1. (B) Linking molecular information from IF (left panel) to the ultrastructural information obtained in EM (middle panel) by overlaying the two imaging modalities based on DAPI and nuclear outlines (dashed lines, right panel). The ultrastructure of the individual LC3-labeled compartments, as exemplified by box 1 (right panel), is shown in C. (C) Ultrastructure of LC3-positive compartments. CLEM images are shown on the left and pseudocolored (beige) EM images on the right (uncolored EM images are shown in Supplemental Figure S1). Inner and outer autophagosomal membranes are indicated by white and black arrowheads, respectively. The autophagic content inside the autolysosome in example 6 is indicated by *. Scale bars = 10 µm (A), 1 µm (B), 200 nm (C). Please click here to view a larger version of this figure.
Supplemental Figure S1: Uncolored EM images of LC3-positive organelles. (A-F) Uncolored EM images of pseudocolored examples 1-6 shown in Figure 2C. The organelles were selected by LC3 fluorescence, as described for Figure 2. Inner and outer autophagosomal membranes are indicated by white and black arrowheads, respectively. The autophagic content inside the autolysosome in example 6 is indicated by *. Scale bars = 200 nm. Abbreviations: AL = autolysosome; AP = autophagosome; M = mitochondrion. Please click here to download this File.
Supplemental File 1: Buffers and solutions used in this study. This supplemental file contains the recipes and protocols needed to make the buffers and solutions used in this study. Please click here to download this File.
Discussion
The method presented here takes advantage of recent advances in cryosection-based on-section CLEM - the high sensitivity of IF labeling and accurate (<100 nm error) correlation between FM and EM14,24. This results in a method with the sensitivity to fluorescently label scarce, endogenous proteins and the capability to overlay this with high precision to the EM ultrastructure. Thus, this method avoids the need for (over)expression of exogenously tagged proteins and the use of less sensitive EM labels. The feasibility of the method is shown by examples of CLEM on endogenous LC3 in starved cells, without the use of lysosomal inhibitors.
Thawed cryosections obtained with the Tokuyasu method are ideal samples for immuno-EM, as unlike resin sections, they are permeable for antibodies. Combined with mild fixation and contrasting procedures, this generally yields superb labeling efficiency over other methods without compromising the detailed ultrastructure, and excellently visualizes cellular membranes12,25,26. Moreover, cryosections are highly compatible with fluorescence microscopy, which make them valuable substrates for CLEM. Both classic immunogold labeling and CLEM on cryosections have provided seminal insights in understanding subcellular organization14,27,28,29,30.
Currently, applications of CLEM on thawed cryosections are becoming more prevalent, as a result of continuous developments and optimizations14,20,24,31,32,33,34 that have improved the quality, applicability, and accuracy of the approach. Now, by accurate correlation of large IF and EM image tilesets, the technique facilitates screening for the ultrastructure of fluorescently-labeled endogenous cellular components14,32,33. This is an advantage over classic immuno-EM, where the search for gold-labeled structures typically requires high magnification and is, therefore, more laborious and time-intensive. It is for this reason that localization of LC3 to the ultrastructure greatly benefits from CLEM. LC3-positive organelles are common when autophagic clearance is blocked (i.e., when cells are treated with BafA1 or pH-raising agents), whereas autophagic organelles are rapidly cleared in unaltered or starved cells, resulting in very low steady-state levels. In such conditions, finding LC3-labeled organelles using classical immuno-EM can be challenging, and CLEM offers a clear advantage.
Previously, CLEM on resin sections was applied in studies using ectopic expression of LC3-GFP or an LC3-GFP-RFP tandem probe35,36,37,38,39. In these studies, fluorescence imaging was performed prior to embedding or directly in acrylic resin sections40, and samples were subsequently screened by EM. There are several advantages of resin embedding; the autophagosomal ultrastructure is generally well-preserved, especially if the material is high-pressure frozen40. Moreover, the contrast of heavy metal-stained resin-embedded material is generally more pronounced than that of uranyl-stained cryosections. Resin-embedded sections are compatible with volumetric EM methods, such as array tomography, FIB-SEM, or serial blockface SEM, while cryosections are not. In approaches that perform imaging before embedding, live-cell imaging is an option41 that is not available in CLEM on cryosections. The key advantage of CLEM on cryosections over these alternatives is the high IF signal, allowing for immuno-localization of rare proteins without the need for membrane permeabilization or overexpression. This avoids potential membrane extraction, overexpression artefacts42 and genetic modification of the subject, which, combined with the possibility to correlate large areas in IF and EM, makes it an excellent tool to study LC3 and autophagy.
Here, the application of on-section CLEM to starved HEPG2 cells revealed that LC3 predominantly localized to structures identified as autophagosomes. Additionally, a few weakly fluorescent spots were found in autolysosomes. This is in direct contrast to cells treated with BafA19 and reflects the rapid degradation of autophagosomal proteins once the autophagosome fuses with lysosomes. Overall, the data demonstrated that CLEM of thawed cryosections can provide insights on LC3-mediated autophagy in native conditions. The data also highlight the sensitivity of the technology, since LC3 was detected even in autolysosomes that contain only low levels of intact LC3 epitopes. Further application of this technique by imaging LC3 in different models and conditions will improve our understanding of autophagy and other LC3-mediated biological processes, such as LC3-associated phagocytosis or conjugation of ATG8 to single membranes.
Beyond autophagy, on-section CLEM can be applied to other rare events or structures, such as cell division, infection, rare cell types in tissues, kinetochores, primary cilia, or cell type-specific organelles. Effective screening for the subject of interest by IF can greatly facilitate the ultrastructural study of these rarities. Furthermore, it was shown14 that the technique can be used to localize proteins in a more sensitive manner than classical immuno-EM. Adjusting the fixation length can further extend this sensitivity, allowing for the ultrastructural localization of very low-abundant or poorly antigenic proteins. Finally, the on-section CLEM method eases rapid selection of a quantitative number of organelles, facilitating a more robust analysis of the ultrastructural distribution of a given protein.
CLEM on cryosections requires the equipment and expertise for cryosectioning. In groups with access to these tools (e.g., cryomicrotomes), the implementation of on-section CLEM is straightforward and only requires the availability of an automated widefield microscope, a setup most labs have access to. Furthermore, the method is available in EM facilities worldwide. Since on-section CLEM combines the application of established IF and EM methods, the method is easily adapted and can be combined with, for example, tomography20,33,43, serial section volume EM of a limited number of sections44, or super-resolution microscopy45. This versatility of the method supports applications to a wide range of biological questions.
Disclosures
The authors declare there are no conflicts of interest.
Acknowledgements
We thank our colleagues at the Center for Molecular Medicine of the University Medical Center Utrecht for fruitful discussions and feedback. We thank past and present colleagues of the Klumperman lab for making continuous improvements in our microscopy technologies. The EM infrastructure used for this work is part of the research program National Roadmap for Large-Scale Research Infrastructure (NEMI) financed by the Dutch Research Council (NWO), project number 184.034.014 to JK.
Materials
| Name | Company | Catalog Number | Comments |
| Chemicals and reagents | |||
| Antibody donkey anti-mouse Alexa Fluor 488 | Life Technologies | #A21202 | use 1:250 |
| Antibody donkey anti-rabbit Alexa Fluor 568 | Life Technologies | A#10042 | use 1:250 |
| Antibody mouse anti-LC3 | Cosmo Bio | CTB-LC3-2-IC | use 1:100 |
| Antibody rabbit anti-LAMP1 | Cell Signaling | 9091 | use 1:250 |
| Bovine serum Albumin, fraction V | Sigma-Aldrich | A-9647 | |
| BSA-c | Aurion | 900.099 | |
| BSA-conjugated gold | Cell Microscopy Core, UMC Utrecht | BSAG 5 nm | |
| Water-free Chloroform | Merck | 1.02447.0500 | |
| DAPI | Invitrogen | 10184322 | Use at end concentration of 10 µg/ml |
| EGTA | Sigma-Aldrich | E4378 | |
| Fish-skin Gelatin | Sigma-Aldrich | G7765 | |
| Food-grade gelatin | Merck | G1890 | |
| Formvar, Vinylec E | SPI | 02492-RA | |
| Gluteraldehyde | Serva | 23115.01 | See CAUTION note |
| Glycerol | Boom | MBAK 7044.1000 | |
| Glycine | Merck | 1042010250 | |
| HEPES | Sigma-Aldrich | H3375 | |
| Methylcellulose, 25 centipoises | Sigma-Aldrich | M-6385 | |
| MgSO4 | Riedel-de Haen | 12142 | |
| Na2HPO4 (PB component A) | Merck | 106580-0500 | |
| NaBH4 | Merck | 806373 | |
| NaH2PO4 (PB component B) | Merck | 106346 | |
| NH4OH | Sigma-Aldrich | 221228-0025 | |
| Oxalic acid | Merck | 100495 | |
| Paraformaldehyde prills | Sigma-Aldrich | 441244 | See CAUTION note |
| PIPES | Merck | 110220 | |
| Protein-A conjugated gold | Cell Microscopy Core, UMC Utrecht | PAG 5, 10, 15 or 20 nm | |
| Sucrose D(+) | VWR | 27483294 | |
| Uranyl acetate | SPI | 020624-AB | See CAUTION note |
| Tools and consumables | |||
| Pick-up loop | Electron Microscopy Sciences | 70944 | |
| Filter paper, qualitative, medium-fast | LLG | 6.242 668 | |
| Finder grids | Ted Pella | G100F1 | |
| Grids | Cell Microscopy Core, UMC Utrecht | CU 100 mesh | |
| Microscopes | |||
| Leica Thunder widefield microscope | Leica | Components: 100x, 1.47 NA TIRF objective; Photometrics prime 95B sCMOS camera; LAS X software; | |
| Leica UC7 ultracryomicrotome | Leica | ||
| Tecnai T12 | FEI | Components: Veleta VEL-FEI-TEC12-TEM camera; SerialEM software | |
| Software | |||
| ec-CLEM in icy | open source | Paul-Gilloteaux et al., 2017 | |
| Fiji | open source | Schindelin et al., 2012 | |
| IMOD | open source | Mastronarde et al., 2017 | |
| Photoshop | Adobe | ||
| SerialEM | open source | Mastronarde et al., 2018 |
References
- Hu, Y., Reggiori, F. Molecular regulation of autophagosome formation. Biochemical Society Transactions. 50 (1), 55-69 (2022).
- Reggiori, F., Ungermann, C. Autophagosome maturation and fusion. Journal of Molecular Biology. 429 (4), 486-496 (2017).
- Kimura, S., Noda, T., Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3 (5), 452-460 (2007).
- Baba, M., Takeshige, K., Baba, N., Ohsumi, Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. The Journal of Cell Biology. 124 (6), 903-913 (1994).
- Takeshige, K., Baba, M., Tsuboi, S., Noda, T., Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. The Journal of Cell Biology. 119 (2), 301-311 (1992).
- De Duve, C., Wattiaux, R. Functions of lysosomes. Annual Review of Physiology. 28, 435-492 (1966).
- Arstila, A. U., Trump, B. F. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. The American Journal of Pathology. 53 (5), 687-733 (1968).
- Eskelinen, E. L., Reggiori, F., Baba, M., Kovács, A. L., Seglen, P. O. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy. 7 (9), 935-956 (2011).
- De Mazière, A., et al. An optimized protocol for immuno-electron microscopy of endogenous LC3. Autophagy. 18 (12), 3004-3022 (2022).
- López-Terrada, D., Cheung, S. W., Finegold, M. J., Knowles, B. B. Hep G2 is a hepatoblastoma-derived cell line. Human Pathology. 40 (10), 1512-1515 (2009).
- Tanida, I., Minematsu-Ikeguchi, N., Ueno, T., Kominami, E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 1 (2), 84-91 (2005).
- Slot, J. W., Geuze, H. J. Cryosectioning and immunolabeling. Nature Protocols. 2 (10), 2480-2491 (2007).
- Waterman-Storer, C. M. Microtubule/organelle motility assays. Current Protocols in Cell Biology. 13 (1), (2001).
- vander Beek, J., de Heus, C., Liv, N., Klumperman, J. Quantitative correlative microscopy reveals the ultrastructural distribution of endogenous endosomal proteins. The Journal of Cell Biology. 221 (1), e202106044(2022).
- Mastronarde, D. N. Advanced data acquisition from electron microscopes with SerialEM. Microscopy and Microanalysis. 24, 864-865 (2018).
- Mastronarde, D. N., Held, S. R. Automated tilt series alignment and tomographic reconstruction in IMOD. Journal of Structural Biology. 197 (2), 102-113 (2017).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Paul-Gilloteaux, P., et al. EC-CLEM: Flexible multidimensional registration software for correlative microscopies. Nature Methods. 14 (2), 102-103 (2017).
- Heiligenstein, X., Paul-Gilloteaux, P., Raposo, G., Salamero, J. eC-CLEM: A multidimension, multimodel software to correlate intermodal images with a focus on light and electron microscopy. Methods in Cell Biology. 140, 335-352 (2017).
- Fermie, J., et al. Bimodal endocytic probe for three-dimensional correlative light and electron microscopy. Cell Reports Methods. 2 (5), 100220(2022).
- Fokkema, J., et al. Fluorescently labelled silica coated gold nanoparticles as fiducial markers for correlative light and electron microscopy. Scientific Reports. 8 (1), 13625(2018).
- Czaja, M. J., et al. Functions of autophagy in normal and diseased liver. Autophagy. 9 (8), 1131(2013).
- Robinson, J. M., Takizawa, T., Pombo, A., Cook, P. R. Correlative fluorescence and electron microscopy on ultrathin cryosections: Bridging the resolution gap. Journal of Histochemistry and Cytochemistry. 49 (7), 803-808 (2001).
- Mohammadian, S., et al. High accuracy, fiducial marker-based image registration of correlative microscopy images. Scientific Reports. 9 (1), 3211(2019).
- Tokuyasu, K. T. A study of positive staining of ultrathin frozen sections. Journal of Ultrasructure Research. 63 (3), 287-307 (1978).
- Slot, J. W., Geuze, H. J. A new method of preparing gold probes for multiple-labeling cytochemistry. European Journal of Cell Biology. 38 (1), 87-93 (1985).
- Klumperman, J., Raposo, G. The complex ultrastructure of the endolysosomal system. Cold Spring Harbor Perspectives in Biology. 6 (10), a016857(2014).
- Geuze, H. J., Slot, J. W., Strous, G. J., Lodish, H. F., Schwartz, A. L. Intracellular site of asialoglycoprotein receptor-ligand uncoupling: Double-label immunoelectron microscopy during receptor-mediated endocytosis. Cell. 32 (1), 277-287 (1983).
- Biazik, J., Ylä-Anttila, P., Vihinen, H., Jokitalo, E., Eskelinen, E. L. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy. 11 (3), 439-451 (2015).
- Fahimi, H. D., Reich, D., Völkl, A., Baumgart, E. Contributions of the immunogold technique to investigation of the biology of peroxisomes. Histochemistry and Cell Biology. 106 (1), 105-114 (1996).
- Vicidomini, G., et al. A novel approach for correlative light electron microscopy analysis. Microscopy Research and Technique. 73 (3), 215-224 (2010).
- Vicidomini, G., et al. High data output and automated 3D correlative light-electron microscopy method. Traffic. 9 (11), 1828-1838 (2008).
- Cortese, K., et al. 3D HDO-CLEM: cellular compartment analysis by correlative light-electron microscopy on cryosection. Methods in Cell Biology. 111, 95-115 (2012).
- van Rijnsoever, C., Oorschot, V., Klumperman, J. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nature Methods. 5 (11), 973-980 (2008).
- Razi, M., Chan, E. Y. W., Tooze, S. A. Early endosomes and endosomal coatomer are required for autophagy. The Journal of Cell Biology. 185 (2), 305-321 (2009).
- Ligeon, L. A., Barois, N., Werkmeister, E., Bongiovanni, A., Lafont, F. Structured illumination microscopy and correlative microscopy to study autophagy. Methods. 75, 61-68 (2015).
- Biazik, J., Vihinen, H., Anwar, T., Jokitalo, E., Eskelinen, E. L. The versatile electron microscope: An ultrastructural overview of autophagy. Methods. 75, 44-53 (2015).
- Gudmundsson, S., Kahlhofer, J., Baylac, N., Kallio, K., Eskelinen, E. L. Correlative light and electron microscopy of autophagosomes. Methods in Molecular Biology. 1880, 199-209 (2019).
- Kriel, J., et al. Correlative light and electron microscopy (CLEM): bringing together the best of both worlds to study neuronal autophagy. Imaging and Quantifying Neuronal Autophagy. 171, 135-147 (2022).
- Largeau, C., Legouis, R. Correlative light and electron microscopy to analyze LC3 proteins in Caenorhabditis elegans embryo. Methods in Molecular Biology. 1880, 281-293 (2019).
- Fermie, J., et al. Single organelle dynamics linked to 3D structure by correlative live-cell imaging and 3D electron microscopy. Traffic. 19 (5), 354-369 (2018).
- Kuma, A., Matsui, M., Mizushima, N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of LC3 localization. Autophagy. 3 (4), 323-328 (2007).
- Ladinsky, M. S., Howell, K. E. Electron tomography of immunolabeled cryosections. Methods in Cell Biology. 79, 543-558 (2007).
- Oorschot, V., Lindsey, B. W., Kaslin, J., Ramm, G. TEM, SEM, and STEM-based immuno-CLEM workflows offer complementary advantages. Scientific Reports. 11 (1), 899(2021).
- Franke, C., et al. Correlative single-molecule localization microscopy and electron tomography reveals endosome nanoscale domains. Traffic. 20 (8), 601-617 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved