Method Article
Whole Genome Sequencing for Rapid Characterization of Rabies Virus Using Nanopore Technology
In This Article
Summary
Here, we present a rapid and cost-effective workflow for characterizing rabies virus (RABV) genomes using nanopore technology. The workflow is intended to support genomics-informed surveillance at a local level, providing information on circulating RABV lineages and their placement within regional phylogenies to guide rabies control measures.
Abstract
Genomic data can be used to track the transmission and geographic spread of infectious diseases. However, the sequencing capacity required for genomic surveillance remains limited in many low- and middle-income countries (LMICs), where dog-mediated rabies and/or rabies transmitted by wildlife such as vampire bats pose major public health and economic concerns. We present here a rapid and affordable sample-to-sequence-to-interpretation workflow using nanopore technology. Protocols for sample collection and the diagnosis of rabies are briefly described, followed by details of the optimized whole genome sequencing workflow, including primer design and optimization for multiplex polymerase chain reaction (PCR), a modified, low-cost sequencing library preparation, sequencing with live and offline base calling, genetic lineage designation, and phylogenetic analysis. Implementation of the workflow is demonstrated, and critical steps are highlighted for local deployment, such as pipeline validation, primer optimization, inclusion of negative controls, and the use of publicly available data and genomic tools (GLUE, MADDOG) for classification and placement within regional and global phylogenies. The turnaround time for the workflow is 2-3 days, and the cost ranges from $25 per sample for a 96 sample run to $80 per sample for a 12 sample run. We conclude that setting up rabies virus genomic surveillance in LMICs is feasible and can support progress toward the global goal of zero dog-mediated human rabies deaths by 2030, as well as enhanced monitoring of wildlife rabies spread. Moreover, the platform can be adapted for other pathogens, helping to build a versatile genomic capacity that contributes to epidemic and pandemic preparedness.
Introduction
The rabies virus (RABV) is a lyssavirus in the Rhabdoviridae family that causes a fatal neurological disease in mammals1. Although rabies is 100% preventable by vaccination, it remains a major public health and economic concern in endemic countries. Of the 60,000 human rabies deaths estimated to occur each year, over 95% are in Africa and Asia where dogs are the primary reservoir2. In contrast, dog vaccination has led to the elimination of dog-mediated rabies across Western Europe, North America, and much of Latin America. In these regions, reservoirs of rabies are now restricted to wildlife, such as bats, raccoons, skunks, and wild canids3. Across Latin America, the common vampire bat is a problematic source of rabies due to regular spillover transmission from bats to both humans and livestock during nightly blood feeding4. The annual global economic impact of rabies is estimated to be $8.6 billion, with livestock losses accounting for 6%5.
Sequence data from viral pathogens combined with metadata on the timing and source of infections can provide robust epidemiological insights6. For RABV, sequencing has been used to investigate the origin of outbreaks7,8, identify host associations with wildlife or domestic dogs8,9,10,11,12, and trace sources of human cases13,14. Outbreak investigations using phylogenetic analysis have indicated that rabies emerged in the formerly rabies-free province of Bali, Indonesia, through a single introduction from the nearby endemic areas of Kalimantan or Sulawesi15. Meanwhile, in the Philippines, an outbreak on Tablas Island, Romblon Province was proven to be introduced from the main island of Luzon16. Viral genomic data have also been used to better understand pathogen transmission dynamics required for targeting control measures geographically. For example, genomic characterization of RABV illustrates the geographic clustering of clades17,18,19, co-circulation of lineages20,21,22, human-mediated viral movement17,23,24, and metapopulation dynamics25,26.
Disease monitoring is one important function of genomic surveillance that has been enhanced with the global increase in sequencing capacity in response to the SARS-CoV-2 pandemic. Genomic surveillance has supported the real-time tracking of SARS-COV-2 variants of concern27,28 and associated countermeasures6. Advances in accessible sequencing technology, such as nanopore technology, have led to improved and more affordable protocols for the rapid sequencing of both human29,30,31,32 and animal33,34,35 pathogens. However, in many rabies endemic countries, there are still barriers to operationalizing pathogen genomic surveillance, as shown by global disparities in SARS-CoV-2 sequencing capacity36. Limitations in laboratory infrastructure, supply chains, and technical knowledge make the establishment and routinization of genomic surveillance challenging. In this paper, we demonstrate how an optimized, rapid, and affordable whole genome sequencing workflow can be deployed for RABV surveillance in resource-limited settings.
Protocol
The study was approved by the Medical Research Coordinating Committee of the National Institute for Medical Research (NIMR/HQ/R.8a/vol.IX/2788), the Ministry of Regional Administration and Local Government (AB.81/288/01), and Ifakara Health Institute Institutional Review Board (IHI/IRB/No:22-2014) in Tanzania; the University of Nairobi Institute of Tropical and Infectious Diseases (P947/11/2019) and the Kenya Medical Research Institute (KEMRI-SERU; protocol No. 3268) in Kenya; and the Research Institute for Tropical Medicine (RITM), Department of Health (2019-023) in the Philippines. Sequencing of samples originating from Nigeria was undertaken on archived diagnostic material collected as a part of national surveillance.
NOTE: Section 1-4 are prerequisites. Section 5-16 describe the sample-to-sequence-to-interpretation workflow for RABV nanopore sequencing (Figure 1). For subsequent steps in the protocol that need pulse centrifugation, centrifuge at 10-15,000 x g for 5-15 s.
1. Computational environment setup for sequencing and data analysis
- Open the Oxford Nanopore Technology (ONT) website37 and create an account to access nanopore-specific resources.
- Log in and install the ONT sequencing and basecalling software38.
- Open GitHub39 and create an account.
- Go to the artic-rabv40 and MADDOG repositories41 and follow the installation instructions.
2. Design or update the multiplex primer scheme
NOTE: Existing RABV schemes are available in the artic-rabv repository40. When targeting a new geographic area, a new scheme should be designed, or an existing scheme modified to incorporate additional diversity.
- Choose a genome reference set to represent the diversity in the study area; this is typically a set of publicly available sequences (e.g., from NCBI GenBank) or preliminary in-house data. Follow step 2.1.1 to use RABV-GLUE42, a RABV sequence data resource, to filter and download NCBI sequences and associated metadata.
NOTE: Choose reference sequences with complete genomes (i.e., without gaps and masked bases). Choosing up to 10 sequences as a reference set for primer design is recommended. If the available sequence data is incomplete or not representative of the study area, refer to the advice43,44,45 in Supplementary File 1.- Navigate to the NCBI RABV Sequences by Clade page from the Sequence Data drop-down menu in RABV-GLUE. Click the Rabies Virus (RABV) link to access all available data or select a particular clade of interest. Use the filter option to Add filters that fit the desired criteria (e.g., country of origin, sequence length). Download sequences and metadata.
- Generate a primer scheme for multiplex polymerase chain reaction (PCR) following the instructions provided by Primal Scheme46. A 400 bp scheme with a 50 bp overlap is recommended to sequence low-quality samples. Download and save all outputs (do not edit the file or primer names).
NOTE: The scheme will be indexed to the first sequence in the input fasta, henceforth referred to as the 'index reference' (Figure 2). See Supplementary File 1 for options to optimize primer performance.
3. Set up RAMPART and ARTIC bioinformatics pipelines
- Refer to Supplementary File 2 to set up a directory structure to manage the input/output files for RAMPART and the ARTIC bioinformatics pipeline.
4. Biosafety and laboratory setup
- Handle potentially rabies-positive samples in biosafety level (BSL) 2 or 3 conditions.
- Ensure laboratory staff have completed rabies pre-exposure vaccination and undergo monitoring of immunity according to World Health Organization (WHO) recommendations3.
- Ensure dedicated standard operating procedures and risk assessments, following national or international guidelines, are in place for the laboratory.
- Required lab setup: Minimize contamination by maintaining physical separation between pre- and post-PCR areas. In laboratories with limited space or in-field lab settings, use portable glove boxes or makeshift lab stations to minimize contamination.
- In this protocol, ensure to designate separate areas for:
- Sample extraction: Set up a BSL2/3 cabinet/glove box to handle biological material and perform inactivation and RNA extraction.
- Template area: Set up a BSL1 cabinet/glove box for the addition of template (RNA/cDNA) to the pre-prepared reaction master mix.
- Master mix area: Set up a designated clean area (BSL1 cabinet/glove box) for the preparation of reagent master mixes. There should be no template in this area.
- Post-PCR area: Set up a separate area for work on amplicons and sequencing library preparation.
NOTE: All areas should be cleaned with a surface decontaminant and ultraviolet (UV)-sterilized before and after use.
5. Field sample collection and diagnosis
NOTE: Samples must be collected by trained and immunized personnel wearing personal protective equipment and following the referenced standard procedures47,48,49.
- Collect the sample via the foramen magnum (i.e., the occipital route), as described in detail in Mauti et al.50.
- Diagnose rabies in the field with rapid diagnostic tests and confirm in the laboratory using recommended procedures47, such as the direct fluorescent antibody test (DFA), the direct rapid immunohistochemical test (DRIT)51,52, or real time reverse transcription (RT)-PCR53.
- Use confirmed positive brain samples for RNA extraction or store in a freezer at -20 °C for 2-3 months or -80 °C for longer periods. Preserve RNA for storage and transport using a suitable DNA/RNA stabilization medium.
6. Sample preparation and RNA extraction (3 h)
NOTE: Use a spin column-based viral RNA extraction kit suitable for the sample type.
- Prepare two ceramic bead tubes by filling a 2 mL PCR tube with approximately 200 µL tube full of 1.4 mm ceramic beads and label the tube.
- Add the recommended volume of lysis buffer provided in the RNA extraction kit to the labeled PCR tube.
- Get approximately a 3 mm cube from the brain sample confirmed with rabies infection using a wooden applicator and put into a labeled tube with sample ID and 100 µL of nuclease-free water into the tube labeled negative control.
NOTE: Use closed tube bead-based homogenization to limit sample exposure. If not possible, use other suitable mechanical disruptors (e.g., rotor-based) or a manual micro pestle. However, these may be less effective than bead beating on hard surface to disrupt tissue (tissue samples may harden in certain storage media). - Disrupt the brain tissue manually using a wooden applicator stick and then vortex at maximum speed until complete tissue homogenization is achieved.
- Centrifuge the lysate as per the manufacturer's instructions and use a pipette to transfer the supernatant to a new labeled microcentrifuge tube. Only use this supernatant in the subsequent steps.
- Follow the RNA extraction kit's spin column instructions to obtain purified RNA.
- Include a negative extraction control (NEC) here and take all the way through to the sequencing stage.
7. cDNA preparation (20 min)
- In the master mix area, prepare a master mix for first strand cDNA synthesis according to the number of samples and controls to be processed (with an excess volume of 10% to ensure adequate reagent; Table 1). A no-template control (NTC) should be included at this stage.
- Label 0.2 mL PCR strip tubes and aliquot 5 µL of the master mix into the tubes.
- Take the prepared tubes to the template area. Add 5 µL of RNA into each labeled tube, including the NEC. Add 5 µL of nuclease-free water (NFW) to the NTC.
- Incubate in a thermal cycler following the conditions mentioned in Table 1.
NOTE: Optional pause point: cDNA can be stored at -20 °C for up to 1 month if necessary, but proceeding to PCR is preferred.
8. Primer pool stock preparation (1 h)
NOTE: This step is only necessary if making new stocks from individual primers, after which pre-prepared stock solutions can be used.
- Prepare a primer pool of 100 µM stock in the master mix area.
- Resuspend the lyophilized primers in 1x tris-EDTA (TE) buffer or NFW at a concentration of 100 µM each. Vortex thoroughly and spin down.
NOTE: In the following steps, individual primers are separated into two primer pools-odd numbered (named Pool A) and even numbered (named Pool B)-to avoid interactions between primers flanking amplicon overlaps. These pools of primers generate overlapping 400 bp amplicons spanning the target genome. - Arrange all the odd numbered primers in a tube rack. Generate a primer pool stock by adding 5 µL from each primer to a 1.5 mL microcentrifuge tube labeled "primer scheme name - Pool A (100 µM)".
- Repeat the process for all the even numbered primers and label as "primer scheme name - Pool B (100 µM)".
- Dilute each primer pool 1:10 in molecular grade water to generate 10 µM primer stocks.
NOTE: Make multiple aliquots of 10 µM primer dilutions and freeze them in case of degradation or contamination.
9. Multiplex PCR (5 h)
- Prepare two PCR master mixes, one for each primer pool in the master mix area.
- Use a final concentration of 0.015 µM per primer. Calculate the required primer pool volume for the PCR reaction (Table 2) using the following formula:
Primer pool volume = number of primers x reaction volume x 0.015/concentration (µM) of primer stock
- Use a final concentration of 0.015 µM per primer. Calculate the required primer pool volume for the PCR reaction (Table 2) using the following formula:
- Aliquot 10 µL each of Pool A master mix and Pool B master mix to labeled PCR strip tubes in the template area. For every sample, add 2.5 µL of cDNA (from step 3) to each of the corresponding labeled primer Pool A and B reactions. Excess cDNA can be stored at -20 °C.
- Mix by gently flicking and pulse centrifuge.
- Incubate the samples with the conditions mentioned in Table 2 on a PCR machine.
NOTE: The program does not include a specific extension step due to the long annealing time of 5 min (required due to the high number of primers) and the short length of the amplicons (400 bp) which is sufficient for the extension.
10. PCR clean up and quantification (3.5 h)
- Perform all work from this point on in the post-PCR area.
- Aliquot solid-phase reversible immobilization (SPRI) beads into microcentrifuge tubes from the main bottle. Store at 4 °C.
- Warm a SPRI bead aliquot to room temperature (RT; ~20 °C) and thoroughly vortex until the beads are fully resuspended in the solution.
- In 1.5 mL tubes, combine the primer Pool A and primer Pool B PCR products for each sample. If necessary, add water to bring the volume to 25 µL.
- Add 25 µL of SPRI beads to each sample (1:1 bead:sample ratio). Mix by pipetting up and down or gently tapping the tube.
- Incubate at RT for 10 min, occasionally inverting or flicking the tubes.
- Place on a magnetic rack until the beads and solution have fully separated. Remove and discard the supernatant, taking care not to disturb the bead pellet.
- Wash twice with 80% ethanol (warmed to RT).
- Add 200 µL of ethanol to the pellet. Wait for 30 s to ensure the beads are washed properly.
- Carefully remove and discard the supernatant, trying not to touch the bead pellet.
- Repeat steps 10.8.1-10.8.2 to wash the pellet a second time.
- Remove all traces of ethanol using a 10 µL tip. Air-dry until trace ethanol has evaporated (with small beads this happens quickly, ~30 s); when this happens, the pellet should go from shiny to matt. Take care not to over-dry (if the pellet is cracking, it is too dry), as this will affect DNA recovery.
- Resuspend the beads in 15 µL of NFW and incubate at RT (off magnetic rack) for 10 min.
- Return to the magnetic rack and transfer the supernatant (cleaned product) to a fresh 1.5 mL tube.
- Prepare a 1:10 dilution of each sample in a separate tube (2 µL of product + 18 µL of NFW).
NOTE: Be very careful at this stage to avoid cross-contamination. Only have one amplicon tube open at a time. Aliquot 18 µL of water into the tubes first (in a clean master mix area). - Measure the DNA concentration of each diluted sample using a highly sensitive and specific fluorometer, as described in protocols.io54,55.
11. Normalization (30 min)
- Use the normalization template (Supplementary File 3) and DNA concentration (ng/µL) of each sample to calculate the volume of diluted (or neat) sample required for 200 fmol of each sample in a total volume of 5 µL.
- Label new PCR tubes and add computed volumes of NFW and sample to obtain normalized DNA.
- Use the computed volume for undiluted (neat) samples if over 5 µL of the diluted sample is required to obtain 200 fmol.
NOTE: Optional pause point: At this point, the cleaned-up PCR product can be stored at 4 °C for up to 1 week or placed at -20 °C for longer-term storage if needed.
12. End-prep and barcoding (1.5 h)
NOTE: The next steps assume the use of specific reagents from nanopore-specific barcoding and ligation sequencing kits (see Table of Materials for details). The protocol is transferable across different chemistry versions, but users should take care to use compatible kits, according to manufacturer instructions.
- End repair and dA-tailing
- Set up the end-prep reaction for each sample mentioned in Table 3. Prepare a master mix according to the number of samples (plus 10% excess). Take care when pipetting as reagents are viscous.
- Add 5 µL of master mix into each tube of normalized DNA (5 µL). The total reaction mix should be 10 µL. Change the tips each time and only have one tube open at a time.
- Incubate in a thermal cycler under the conditions mentioned in Table 3.
- Barcoding
- Aliquot the barcodes from the barcoding kit to PCR strip tubes at 1.25 µL/tube, and record barcode assigned to each sample.
- Add 0.75 µL of the end-prepped sample to its assigned barcode aliquot.
- Prepare a ligation master mix according to the number of samples (plus 10% excess) (Table 4).
- Add 8 µL of ligation master mix to the end-prepped sample + barcodes, giving a total reaction of 10 µL.
- Incubate in a thermal cycler using the conditions mentioned in Table 4.
- SPRI bead clean up and DNA quantification
- Thaw short fragment buffer (SFB) at RT, mix by vortexing, pulse centrifuge, and place on ice.
- Pool all the barcoded samples together in a 1.5 mL lobind microcentrifuge tube. So as not to make the clean up volume too large to use, pool 12-24 samples (10 µL/sample), up to 48 samples (5 µL/sample), or up to 96 samples (2.5 µL/sample) from each native barcoding reaction.
- Add a 0.4x volume of SPRI beads to the barcoded pool. Mix gently (flicking or pipetting) and incubate at RT for 5 min.
- Place the samples on a magnet until the beads have pelleted and the supernatant is completely clear (~2 min). Remove and discard the supernatant. Take care not to disturb the beads.
- Wash twice with 250 µL of SFB.
- Remove the tube from the magnet and completely resuspend the pellet in 250 µL of SFB. Incubate for 30 s, pulse centrifuge, and return to the magnet.
- Remove the supernatant and discard.
- Repeat step 12.3.5 to perform a second SFB wash.
- Pulse centrifuge and remove any residual SFB.
- Add 200 µL of 80% (RT) ethanol to bathe the pellet. Remove and discard the ethanol, being careful not to disturb the bead pellet. Air-dry for 30 s or until the pellet has lost its shine.
- Resuspend in 22 µL of NFW at RT for 10 min.
- Place on the magnet, leave to settle for ~2 min, then carefully remove the solution and transfer to a clean 1.5 mL microcentrifuge tube.
- Use 1 µL to obtain the DNA concentration, as described previously (step 10.13).
NOTE: Optional pause point: At this point, the library can be stored at 4 °C for up to 1 week or -20 °C for longer-term storage, but it is preferable to continue with adapter ligation and sequencing.
13. Sequencing (48 h maximum)
- Prepare the computer (refer also to Prerequisites sections 1-4).
- Check that there is enough space to store new data (min 150 GB), data from old runs are backed up/moved to a server before deleting, and the latest version of MinKNOW is installed.
- Remove the stored flow cell from the fridge and allow it to reach RT.
- Adapter ligation (1 h)
- Pulse centrifuge the adapter mix and ligase and place on ice.
- Thaw elution buffer (EB), SFB, and ligation buffer at RT. Mix by vortexing, pulse centrifuge, and place on ice.
- Prepare the adapter ligation master mix (Table 5), combining reagents in the specified order in a low bind tube.
NOTE: Alternatives for adapter ligation master mix reagents (Table 5) can be used depending on availability at the lab. See Supplementary File 3 and Table of Materials for a list of alternatives. Use computation in the Supplementary File 3 worksheet to get the volume of DNA library equivalent to 200 fmol. If less than 20 µL is computed, add NFW to make up to 20 µL. - Mix by gentle flicking and pulse centrifuge. Incubate at RT for 20 min.
NOTE: During incubation, start preparing the flow cell (section 13.5).
- Clean up using SPRI beads (do not use ethanol as in earlier clean ups).
- Add a 0.4x volume of SPRI beads (RT) to the samples. Incubate at RT for 10 min, gently flick intermittently to aid mixing.
- Place on the magnet until the beads and solution have fully separated (~5 min). Remove and discard the supernatant; take care not to disturb the bead pellet.
- Wash twice with 125 µL of SFB.
- Resuspend the pellet completely with 125 µL of SFB by mixing with a pipette. Leave to incubate for 30 s.
- Pulse centrifuge to collect liquid at the tube base and place on the magnet. Remove the supernatant and discard.
- Repeat steps 13.4.4-13.4.5 to wash the pellet a second time.
- Pulse centrifuge and remove excess SFB.
- Resuspend in 15 µL of EB and incubate for 10 min at RT.
- Return to the magnet for ~2 min and then carefully transfer the solution to a clean 1.5 mL microcentrifuge tube.
- Quantify 1 µL of the eluted library, as described previously in step 10.13
NOTE: For best results, proceed directly to MinION sequencing; however, the final library can be stored in EB at 4 °C for up to 1 week if needed.
- Run a flow cell quality check.
- Connect the sequencing device to a laptop and open the sequencing software.
- Select flow cell type, and then click Check Flow Cell and Start Test.
- Once complete, the total number of active (i.e., viable) pores will be displayed. A new flow cell should have >800 active pores; if it does not, contact the manufacturer for a replacement.
- Priming and loading the flow cell (20 min)
- Thaw the following reagents at RT and then place sequencing buffer, a flush tether, flush buffer, and loading beads on ice.
- Vortex the sequencing buffer and flush buffer, pulse centrifuge, and place on ice.
- Pulse centrifuge the flush tether and mix by pipetting; then place on ice.
- Prepare the flow cell priming mix by adding 30 µL of flush tether directly to the tube of flush buffer from a flow cell priming kit and mix by pipetting.
- Mix the loading beads by pipetting immediately prior to use as they settle quickly.
- In a fresh tube, prepare the final library dilution for sequencing, as mentioned in Table 5.
NOTE: Use computation in the Supplementary File 3 worksheet to get the volume of DNA library equivalent to 50 fmol. If less than 12 µL is computed, add EB to make up to 12 µL. - Flip back the sequencing device lid and slide the priming port cover clockwise so that the priming port is visible (Figure 3)
- Remove air bubbles carefully by setting a P1000 pipette to 200 µL, insert the tip into the priming port, and turn the wheel until a small volume entering the pipette tip is seen (max turn to 230 µL).
- Load 800 µL of flow cell priming mix into the flow cell via the priming port, taking care to avoid bubbles.
- Leave for 5 min.
- Lift the sample port cover gently and load 200 µL of priming mix into the flow cell via the priming port using a P1000 pipette.
- Pipette the library mix up and down prior to loading, ensuring loading beads in the master mix are resuspended before loading.
- Load 75 µL of library mix to the flow cell via the sample port in a dropwise fashion. Ensure that each drip flows into the port before adding the next.
- Replace the sample port cover gently, making sure the bung enters sample port.
- Close the priming port and replace the sequencing device lid.
- Sequencing run (48 h maximum)
- Connect the sequencing device to the laptop and open the sequencing software.
- Click start and then click Start Sequencing.
- Click New Experiment and follow the sequencing software graphical user interface (GUI) workflow to set up the parameters for the run.
- Type in the experiment name and sample ID (e.g., rabv_run1), and choose the Flow Cell Type from the drop-down menu.
- Continue to kit selection and choose the relevant ligation sequencing kit and native barcoding kit(s) used.
- Continue to Run options. Keep the defaults, unless it is desired for the run to stop automatically after a certain number of hours (runs can be stopped manually at any time).
- Continue to Basecalling. Choose to turn Basecalling On or Off according to the computing resources (see computer setup). Choose Edit Options under barcoding and ensure Barcode Both Ends is turned on. Save and continue to the output section.
- Accept the defaults and continue to final review, check the settings, and record the details in worksheet (Supplementary File 3). Click Start.
NOTE: If the flow cell is being reused, adjust the starting voltage (in the advanced section of the run options), as indicated by the scheme in Supplementary File 3. - Record the initial active channels-if this is significantly lower than the quality control (QC) check, restart the sequencing software. If still lower, then reboot the computer.
- Record the initial channels in strand versus single pore to give an approximate pore occupancy. This number will fluctuate, so give an approximation.
- Monitor the run as it progresses.
14. Live and offline basecalling
NOTE: These instructions assume that the pre-existing directory structure provided in the artic-rabv repository and that Prerequisites sections 1 and 3 of the protocol have been followed.
- On your local file system, create a new directory called analysis, where you will store all of your analysis outputs. To organize further: create a sub-directory with the name of your project and inside that a new directory for the run, using the sample ID provided to the MinKNOW as the run_name. Do this in one command as follows:
mkdir -p
analysis/project_name/run_name
Then navigate to its location:
cd
path/analysis/project_name/run_name - Live basecalling
NOTE: To perform nanopore basecalling in real time, laptops require a NVIDIA CUDA-compatible graphics processing unit (GPU). Ensure instructions for the GPU basecalling setup have been performed using the guppy protocol56.- During run setup, turn live basecalling on.
- Use RAMPART to monitor the sequencing coverage in real time, as per the instruction below.
- In the computer's terminal, activate the artic-rabv conda environment:
conda activate artic-rabv - Create a new directory for the rampart output inside the run_name directory and navigate into it:
cd /path/analysis/project_name/run_name
mkdir rampart_output
cd rampart_output - Create a barcodes.csv file to pair the barcodes and sample names. It should have one line per barcode and only specify barcodes that are present in the library, with the headings "barcode" and "sample". Follow the example in the artic-rabv directory:
analysis/example_project/example_run/rampart_output/barcodes.csv - Start RAMPART by providing the relevant protocol folder and path to the fastq_pass folder in the MinKNOW output for the run:
rampart --protocol /path/rampart/scheme_name_V1_protocol - basecalledPath <insertpathTo Fastqpassfolder> - Open a browser window and navigate to localhost:3000 in the URL box. Wait for sufficient data to be basecalled before results appear on the screen.
- Offline basecalling (performed post-run)
- If live basecalling was not set, the output from MinKNOW will be raw signal data (fast5 files). One will not be able to use RAMPART during the run. Convert the fast5 files to basecalled data (fastq files) post-run using guppy (see setup in Prerequisites step 1.1.1.). Run RAMPART post-hoc on the basecalled data.
- Run the guppy basecaller:
guppy_basecaller -c dna_r9.4.1_450bps_fast.cfg -i /path/to/reads/fast5_* -s /path/analysis/project_name/run_name -x auto -r
-c is the config file to specify the basecalling model, -i is the input path, -s is the save path, -x specifies basecalling by the GPU device (exclude if using the computer version of guppy), and -r specifies to search input files recursively.
NOTE: The config file (.cfg) can be changed to a high-accuracy basecaller by replacing _fast with _hac, although this will take significantly longer.
15. Washing the flow cells
- The flow cells can be washed and reused to sequence new libraries if pores are still viable. See instructions for washing at the ONT flow cell wash protocol57.
16. Analysis and interpretation
- Consensus sequence generation with ARTIC bioinformatics pipeline
- Follow the instructions detailed in the artic-rabv GitHub repository40 in the rabv_protocols folder to generate consensus sequences from raw fast5 or basecalled fastq files.
NOTE: Refer to Artic pipeline - Core pipeline58 for further guidance.
- Follow the instructions detailed in the artic-rabv GitHub repository40 in the rabv_protocols folder to generate consensus sequences from raw fast5 or basecalled fastq files.
- Optional: Analyze the average read depth per amplicon.
- Adapt the scripts available from the artic-rabv repository, referring to Supplementary File 1. Briefly, in-depth statistics are generated using SAMtools59 and coverage per nucleotide plotted in R.
- Phylogenetic analysis using GLUE
- From RABV_GLUE42, select Analysis > Genotyping and Interpretation tab > Add Files, selecting the fasta file of consensus sequences.
- Click Submit and wait. Once the analyses are complete, the Show Analysis button will be available to click, showing clade and subclade assignments, coverage per gene, variation from reference sequences, and closest relative.
- Relevant contextual sequences can also be identified in the Sequence Data > NCBI Sequences by Clade section.
- Select the clade identified or click Rabies Virus (RABV) to see all the available sequences.
- Filter for relevant sequences (e.g., country of origin).
- Download these sequences and corresponding metadata for analysis and comparison.
- Lineage assignment using MADDOG41
- Pull the MADDOG repository from GitHub to ensure you are working with the most up-to-date version.
- Create an assignment folder within the local MADDOG repository (previously created in the Prerequisites section) called the run name.
- Inside the folder, add the fasta file containing the consensus sequences.
- Add a metadata file to the folder.
NOTE: This file must be a csv with 4 columns called 'ID,' 'country,' 'year,' and 'assignment,' detailing the sequence IDs, the country of sampling, and the year of sample collection, while the 'assignment' column should be blank. - In the command line interface, activate the conda environment: conda activate MADDOG.
- In the command line interface, navigate to the MADDOG repository folder.
- Initially, undertake lineage assignment on sequences to check for any potential abnormalities and to identify if running the longer lineage designation step would be appropriate. For this, type this in the command line: sh assignment.sh.
- When prompted, enter Y to indicate you have pulled the repository and are working with the most up-to-date version of MADDOG.
- When prompted, enter the name of the folder within the MADDOG repository folder that contains the fasta file.
- When the lineage assignment is complete, check the output file in your folder. If the output is as expected and there are multiple sequences assigned to the same lineage, run the lineage designation.
- If running lineage designation, delete the assignment output file just created.
- In the terminal, inside the MADDOG repository folder, run the command sh designation.sh.
- When prompted enter Y to indicate you have pulled the repository and are working with the most up-to-date version of MADDOG.
- When prompted, enter the folder name within the MADDOG repository folder containing the fasta file and metadata. This outputs lineage information about each sequence, a phylogeny of the new and relevant previous sequences (from 16.3.6), hierarchical information about the lineages, and details of potentially emerging lineages and areas of undersampling.
NOTE: Full details of the protocol, usage, and outputs can be found in Campbell et al.60. - When the initial analysis has been completed, if prompted to also test for emerging and undersampled lineages, enter Y, if this is required. Otherwise, enter N.
- If prompted to confirm newly found lineages, enter Y and follow instructions in the resultant NEXT_STEPS.eml file. Otherwise, enter N.
Results
The sample-to-sequence-to-interpretation workflow for RABV described in this protocol has been used successfully in different laboratory conditions in endemic countries, such as Tanzania, Kenya, Nigeria, and the Philippines (Figure 4). The protocol was used on different sample types and conditions (Table 6): fresh and frozen brain tissue, cDNA and RNA extracts from brain tissue transported under cold chain for extended periods, and FTA cards with brain tissue smears.
Live basecalling using RAMPART (Figure 5) shows the almost real-time generation of reads and the percent coverage per sample. This is particularly useful in deciding when to stop the run and save the flow cell for reuse. Variation in run time was observed, with some finished in 2 h, while others could take more than 12 h for an adequate depth of coverage (x100) to be reached. We can also view regions with poor amplification; for example, Figure 6 shows a snapshot of one sequencing run where coverage profiles show some amplicons with very low amplification, indicating potentially problematic primers. By investigating these poorly amplifying regions more thoroughly, we have been able to identify primer mismatches, which will enable us to redesign and improve individual primers. Some primer schemes have shown more mismatches than others. This is observed in the East Africa primer scheme, as compared to the Philippines, in line with the targeted diversity, as the East Africa scheme aims to capture a much broader diversity.
RABV-GLUE42, a general-purpose resource for RABV genome data management, and MADDOG60, a lineage classification and nomenclature system, were used to compile and interpret resulting RABV sequences. Table 7 shows the major and minor clades circulating in each country assigned using RABV-GLUE. Also shown is a higher resolution classification of local lineages following the MADDOG assignment.
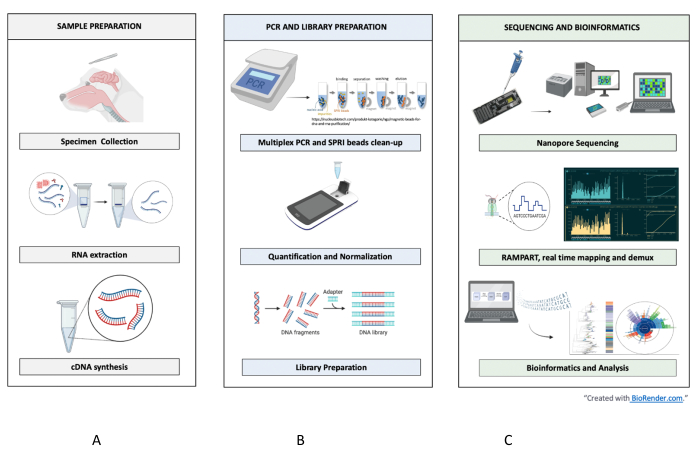
Figure 1: Sample-to-sequence-to-interpretation workflow for RABV. Summarized steps are shown for (A) sample preparation, (B) PCR and library preparation, and (C) sequencing and bioinformatics up to analysis and interpretation. Please click here to view a larger version of this figure.
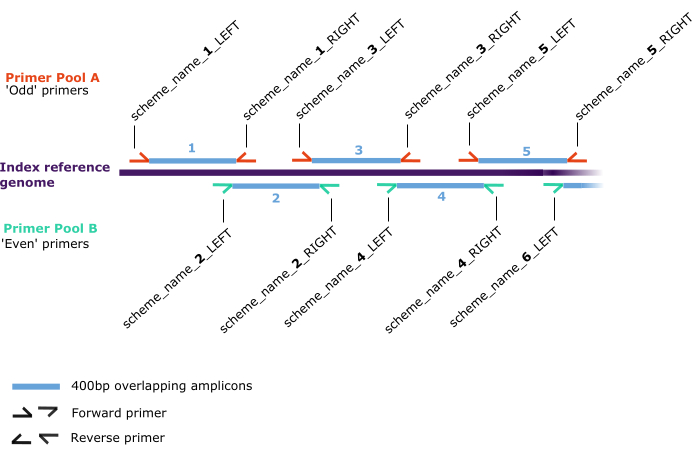
Figure 2: Primer scheme schematic. Annealing positions along the 'index reference genome' (dark purple) for pairs of forward and reverse primers (half arrows), which are assigned in two separate pools: A (red) and B (green). Primer pairs generate 400 bp overlapping amplicons (blue) which are numbered sequentially along the index reference genome in the format 'scheme_name_X_DIRECTION', where 'X' is a number referring to the amplicon generated by the primer and 'DIRECTION' is either 'LEFT' or 'RIGHT', describing the forward or reverse respectively. Odd or even values of 'X' determine the pool (A or B, respectively). Please click here to view a larger version of this figure.
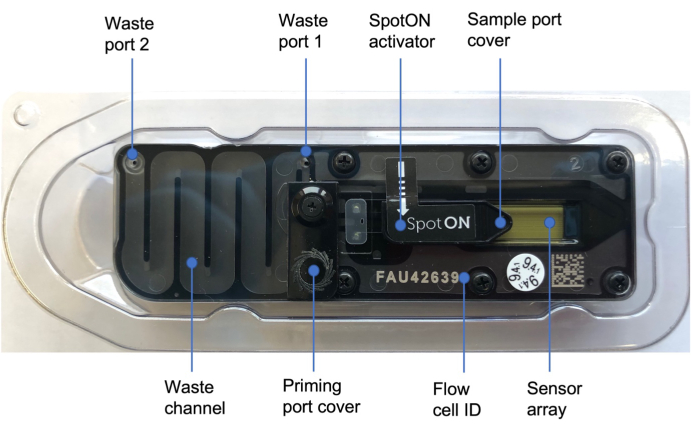
Figure 3: Nanopore flow cell48. Blue labels illustrate the different parts of the flow cell, including the priming port cover which covers the priming port where the priming solution is added, the SpotON sample port cover covering the sample port where the sample is added in a dropwise fashion, the waste ports 1 and 2, and the flow cell ID. Please click here to view a larger version of this figure.
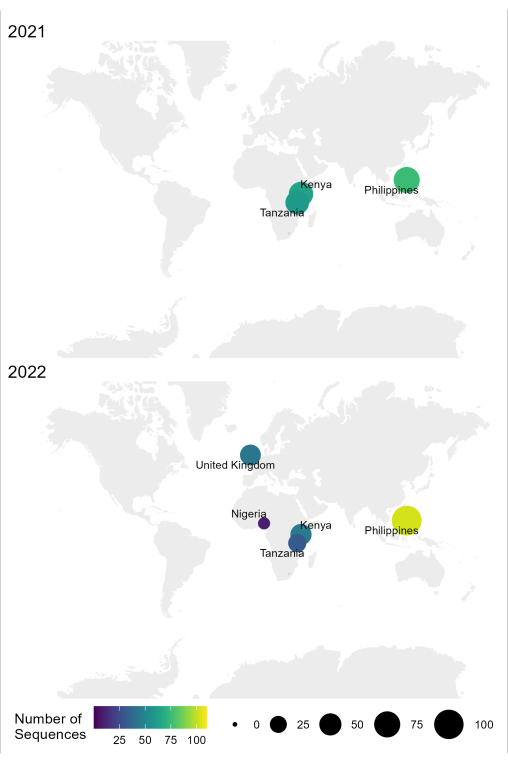
Figure 4: Map showing the location where RABV sequencing was conducted using the optimized workflow in 2021 and 2022. Bubble size and color correspond to the number of sequences per location, where smaller and darker is fewer, while larger and lighter is more. Please click here to view a larger version of this figure.
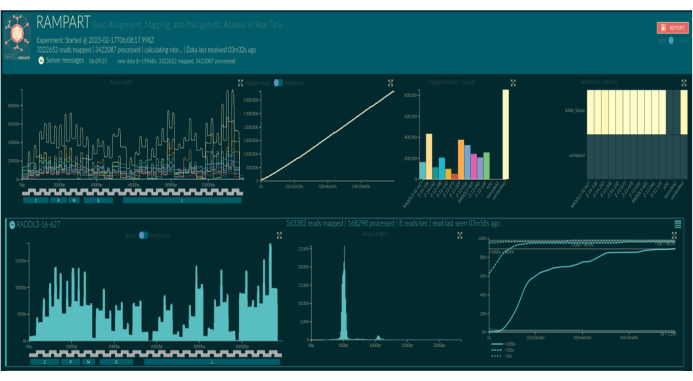
Figure 5: Screenshot of RAMPART visualization in web browser. Barcode names are replaced by sample names according to the bioinformatic setup. The top three panels show summary plots for the whole run: depth of coverage of mapped reads for each barcode per nucleotide position on the index reference genome (top left, colored by barcode), summed mapped reads from all barcodes over time (top middle), and mapped reads per barcode (top right, colored by barcode). Lower panels show rows of plots per barcoded. From left to right: the depth of coverage of mapped reads per nucleotide position on the index reference genome (left), length distribution of mapped reads (middle), and proportion of nucleotide positions on the index reference genome which have obtained a 10x, 100x and 1,000x coverage of mapped reads over time (right). Please click here to view a larger version of this figure.
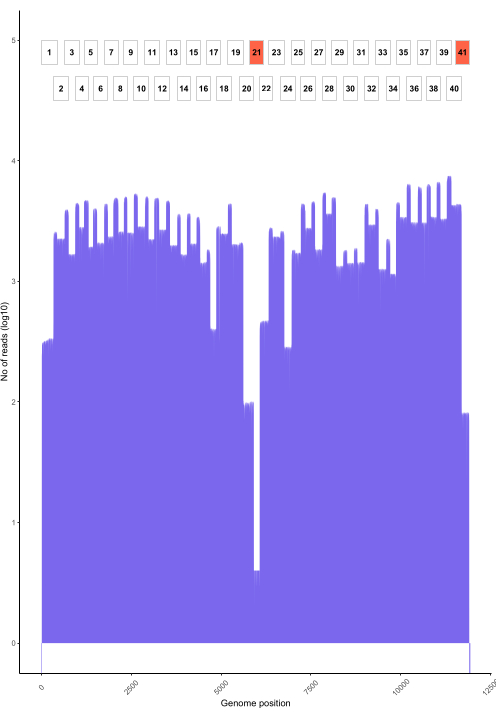
Figure 6: An example read coverage across the genome for a rabies virus sample from the Philippines sequenced using the protocol. Read coverage at each nucleotide position in the genome is shown, alongside the position of the overlapping amplicons (1-41) used to generate the library. Spikes in the depth of coverage correspond to areas of amplicon overlap. Amplicons with a low depth of coverage correspond to areas of amplicon overlap. Amplicons with a low depth of coverage are highlighted in red indicating problematic regions that may require optimization. Please click here to view a larger version of this figure.
Table 1: Master mix and thermal cycler conditions for cDNA preparation. Please click here to download this Table.
Table 2: Master mix and thermal cycler conditions for multiplex PCR. Please click here to download this Table.
Table 3: Master mix and thermal cycler conditions for end-prep reaction. Please click here to download this Table.
Table 4: Master mix and thermal cycler conditions for barcoding. Please click here to download this Table.
Table 5: Adapter ligation master mix and final library dilution for sequencing. Please click here to download this Table.
Table 6: The number of rabies virus whole genome sequences generated and the type of samples used in different countries using the sample-to-sequence-to-interpretation workflow. Please click here to download this Table.
Table 7: Major and minor clade assignments from RABV-GLUE and lineage assignments from MADDOG for sequences generated using the workflow. Please click here to download this Table.
Supplementary File 1: Primer scheme design and optimization, and amplicon read depth analysis. Please click here to download this File.
Supplementary File 2: Computational setup Please click here to download this File.
Supplementary File 3: RABV WGS protocol worksheet Please click here to download this File.
Discussion
An accessible RABV, nanopore-based, whole genome sequencing workflow was developed by Brunker et al.61, using resources from the ARTIC network46. Here, we present an updated workflow, with complete sample-to-sequence-to-interpretation steps. The workflow details the preparation of brain tissue samples for whole genome sequencing, presents a bioinformatics pipeline to process reads and generate consensus sequences, and highlights two rabies-specific tools to automate lineage assignment and determine phylogenetic context. The updated workflow also provides comprehensive instructions for the setup of appropriate computational and laboratory workspaces, with considerations for implementation in different contexts (including low-resource settings). We have demonstrated the successful implementation of the workflow in both academic and research institute settings in four RABV endemic LMICs with no or limited genomic surveillance capacity. The workflow has proven resilient to application across diverse settings, and comprehensible by users with varying expertise.
This workflow for RABV sequencing is the most comprehensive publicly available protocol (covering sample-to-sequence-to-interpretation steps) and specifically adapted to reduce both startup and running costs. The time and cost required for library preparation and sequencing with nanopore technology is greatly reduced relative to other platforms, such as Illumina61, and continual technology developments are improving sequence quality and accuracy to be comparable with Illumina62.
This protocol is designed to be resilient in diverse low-resource contexts. By referring to the troubleshooting and modifications guidance provided alongside the core protocol, users are supported to adapt the workflow to their needs. The addition of user-friendly bioinformatic tools to the workflow constitutes a major development to the original protocol, providing rapid and standardized methods that can be applied by users with minimal prior bioinformatics experience to interpret sequence data in local contexts. The capacity to do this in situ is often limited by the need to have specific programming and phylogenetic skills, which require an intensive and long-term skills training investment. While this skillset is important to thoroughly interpret sequence data, basic and accessible interpretation tools are equally desirable in order to capacitate local "sequencing champions", whose core expertise may be wet lab based, enabling them to interpret and take ownership over their data.
As the protocol has been undertaken for a number of years in several countries, we now can provide guidance on how to optimize multiplex primer schemes to improve coverage and deal with accumulated diversity. Efforts have also been made to help users improve the cost-effectiveness or to allow for ease of procurement in a given region, which is typically a challenge for the sustainability of molecular approaches63. For example, in Africa (Tanzania, Kenya, and Nigeria), we opted for blunt/TA ligase master mix at the adapter ligation step, which was more readily available from local suppliers and a cheaper alternative to other ligation reagents.
From experience, there are several ways of reducing the cost per sample and per run. Reducing the number of samples per run (e.g., from 24 down to 12 samples) can extend the life of flow cells over multiple runs, whereas increasing the number of samples per run maximizes the time and reagents. In our hands, we were able to wash and reuse flow cells for one in every three sequencing runs, enabling an additional 55 samples to be sequenced. Washing the flow cell immediately after use, or if not possible, removing the waste fluid from the waste channel after every run, seemed to preserve the number of pores available for a second run. Taking into consideration the initial number of pores available in a flow cell, one run can also be optimized to plan how many samples to run in a particular flow cell.
Though the workflow aims to be as comprehensive as possible, with the addition of detailed guidance and signposted resources, the procedure is still complex and can be daunting for a new user. The user is encouraged to seek in-person training and support, ideally locally, or alternatively through external collaborators. In the Philippines for example, a project on capacity building within regional laboratories for SARS-CoV-2 genomic surveillance using ONT has developed core competencies among health care diagnosticians that are readily transferrable to RABV sequencing. Important steps, such as SPRI bead clean up, can be difficult to master without hands-on training, and ineffective clean up can damage the flow cell and compromise the run. Sample contamination is always a major concern when amplicons are being processed in the lab and can be difficult to eliminate. In particular, cross-contamination between samples is extremely difficult to detect during post-run bioinformatics. Good laboratory technique and practices, such as maintaining clean work surfaces, separating pre- and post-PCR areas, and incorporating negative controls, are imperative to ensure quality control. The fast pace of nanopore sequencing developments is both an advantage and disadvantage for routine RABV genomic surveillance. Continuing improvements to nanopore's accuracy, accessibility, and protocol repertoire widen and improve the scope for its application. However, the same developments make it challenging to maintain standard operating procedures and bioinformatic pipelines. In this protocol, we provide a document assisting the transition from older to current nanopore library preparation kits (Table of Materials).
A common roadblock to sequencing in LMICs is accessibility, including not only the cost but also the ability to procure consumables in a timely manner (in particular sequencing reagents, which are relatively new to procurement teams and suppliers) and computational resources, as well as simply having access to stable power and the internet. Using portable nanopore sequencing technology as the foundation of this workflow helps with many of these accessibility issues, and we have demonstrated the use of our protocol across a range of settings, conducting the full protocol and analysis in-country. Admittedly, procuring equipment and sequencing consumables in a timely manner remains a challenge and, in many instances, we were forced to carry or ship reagents from the UK. However, in some areas, we were able to rely entirely on local supply routes for reagents, benefiting from investment in SARS-CoV-2 sequencing (e.g., the Philippines) that has streamlined procurement processes and begun to normalize the application of pathogen genomics.
The need for a stable internet connection is minimized by one-time-only installs; for instance, GitHub repositories, software download, and nanopore sequencing itself only require internet access to start the run (not throughout) or can be performed completely offline with agreement from the company. If mobile data is available, a phone can be used as a hotspot to the laptop to begin the sequencing run, before disconnecting for the run duration. When routinely processing samples, data storage requirements can grow rapidly, and ideally data would be stored on a server. Otherwise, solid state drive (SSD) hard drives are relatively cheap to source.
While we recognize that there are still barriers to genomic surveillance in LMICs, increasing investment in building genomics accessibility and expertise (e.g., Africa Pathogen Genomics Initiative [Africa PGI])64 suggests that this situation will improve. Genomic surveillance is critical for pandemic preparedness6, and capacity can be established through routinizing the genomic surveillance of endemic pathogens such as RABV. Global disparities in sequencing capacities highlighted during the SARS-CoV-2 pandemic should be a driver of catalytic change to address these structural inequities.
This sample-to-sequence-to-interpretation workflow for RABV, including accessible bioinformatics tools, has the potential to be used to guide control measures targeting the goal of zero human deaths from dog-mediated rabies by 2030, and ultimately for the elimination of RABV variants. Combined with relevant metadata, genomic data generated from this protocol facilitates rapid RABV characterization during outbreak investigations and in the identification of circulating lineages in a country or region60,61,65. We illustrate our pipeline mostly using examples from dog-mediated rabies; however, the workflow is directly applicable to wildlife rabies. This transferability and low cost minimize the challenges in making routine sequencing easily available, not only for rabies but also for other pathogens46,66,67, to improve disease management and control.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by Wellcome [207569/Z/17/Z, 224670/Z/21/Z], Newton funding from the Medical Research Council [MR/R025649/1] and the Philippines Department of Science and Technology (DOST), the UK Research and Innovation Global effort on COVID-19 [MR/V035444/1], the University of Glasgow Institutional Strategic Support Fund [204820], Medical Research Council New Investigator Award (KB) [MR/X002047/1], and International Partnership Development Fund, a DOST British Council-Philippines studentship (CB), a National Institute for Health Research [17/63/82] GemVi scholarship (GJ), and University of Glasgow studentships from the MVLS DTP (KC) [125638-06], the EPSRC DTP (RD) [EP/T517896/1], and the Wellcome IIB DTP (HF) [218518/Z/19/Z]. We are grateful to colleagues and collaborators who have supported this work: Daniel Streicker, Alice Broos, Elizabeth Miranda, DVM, Daria Manalo, DVM, Thumbi Mwangi, Kennedy Lushasi, Charles Kayuki, Jude Karlo Bolivar, Jeromir Bondoc, Esteven Balbin, Ronnel Tongohan, Agatha Ukande, Davis Kuchaka, Mumbua Mutunga, Lwitiko Sikana, and Anna Czupryna.
Materials
| Name | Company | Catalog Number | Comments |
| Brand name | |||
| Software | |||
| Sequencing software (MinKnow) | Oxford Nanopore Technologies | https://community.nanoporetech.com/downloads | |
| Bioinformatics tool kit (Guppy) | Oxford Nanopore Technologies | https://community.nanoporetech.com/docs/prepare/library_prep_protocols/Guppy-protocol/v/gpb_2003_v1_revao_14dec2018 | |
| Equipment | |||
| Thermal cycler (miniPCR™ mini16 thermal cycler) | Cambio | MP-QP-1016-01 | |
| Homogenizer (Precellys Evolution Touch Homogenizer) | Bertin Instruments | EQ02520-300 | |
| Cold Racks (0.2-0.5mL) (PCR Mini-cooler with transparent lid) | BRAND | 781260 | |
| Pipettor | |||
| (Pipetman L Fixed F1000L, 1000 uL) | Gilson | SKU: FA10030 | |
| (Pipetman L Fixed F100L, 100 uL) | Gilson | SKU: FA10024 | |
| (Pipetman L Fixed F10L, 10 uL) | Gilson | SKU: FA10020 | |
| (Pipetman L Fixed F1L, 1 uL) | Gilson | SKU: FA10025 | |
| (Pipetman L Fixed F20L, 20 uL) | Gilson | SKU: FA10021 | |
| (Pipetman L Fixed F250L, 250 uL) | Gilson | SKU: FA10026 | |
| Fluorometer (Qubit 4 Fluorometer) | Thermofisher scientific/Fisher scientific | Q33238 | |
| Laptop (Any brand with ~2 GB of drive space, minimum of 512 GB storage space, msi installer [GPU]) | |||
| Microcentrifuge (Refrigerated centrifuge) | Thermofisher scientific/Fisher scientific | 75004081 | |
| Vortex mixer (Basic vortex mixer) | Thermofisher scientific/Fisher scientific | 88882011 | |
| Magnetic rack (DynaMag -2 Magnet) | Thermofisher scientific/Fisher scientific | 12321D | |
| Sequencing device (MinION) | Oxford Nanopore Technologies | MinION Mk1B | |
| RNA Extraction | |||
| RNA extraction kit (Qiagen RNEasy Mini Kit 250) | Qiagen | 74106 | |
| RNA stabilizing reagents | |||
| (RNA later) | Invitrogen | AM7020 | |
| (DNA/RNA Shield) | Zymo Research | R1100-50 | |
| PCR | |||
| Nuclease-free Water (Nuclease-free Water [not DEPC-treated]) | Thermofisher scientific/Fisher scientific | AM9937 | |
| Master mix for first strand cDNA synthesis (LunaScript RT SuperMix Kit) | New England Biolabs | E3010S | |
| DNA amplification master mix (Q5® Hot Start High-Fidelity 2X Master Mix [NEB]) | New England Biolabs | M0494L | |
| Primer (Scheme) (Custom DNA oligos) | Invitrogen | ||
| SPRI Bead Clean-up | |||
| SPRI beads (Aline Biosciences PCR Clean DX ) | Cambio | AL-AC1003-50 | |
| Ethanol, Pure Absolute, >99.8% (GC) [Riedel-De Haen] | Merck | 818760 | |
| Short Fragment buffer (SFB expansion pack) | Oxford Nanopore Technologies | EXP-SFB001 | |
| DNA Quantification | |||
| DNA quantification kit (Qubit® dsDNA HS Assay Kit) | Thermofisher scientific/Fisher scientific | Q32854 | |
| DNA quantification assay tubes (Qubit™ Assay Tubes) | Thermofisher scientific/Fisher scientific | Q32856 | |
| End Prep and barcoding (Qubit™ Assay Tubes) | |||
| End Prep master mix (NEBNext Ultra End Repair/dA-Tailing Module) | New England Biolabs | E7546L | |
| Barcoding kit | |||
| *Chemistry 9 | Oxford Nanopore Technologies | *Chemistry 9 | |
| (Native Barcoding Expansion 1-12) | EXP-NBD104 | ||
| (Native Barcoding Expansion 13-24) | EXP-NBD114 | ||
| (Native Barcoding Expansion 96) | EXP-NBD196 | ||
| *Chemistry 14 | Oxford Nanopore Technologies | *Chemistry 14 | |
| (not compatible) | (not compatible) | ||
| (Native Barcoding Kit 24 V14) | SQK-NBD114.24 | ||
| (Native Barcoding Kit 96 V14) | SQK-NBD114.96 | ||
| Ligation mastermix (Blunt/TA Ligase Master Mix) | New England Biolabs | M0367S | |
| Adapter Ligation | |||
| Adapter ligation master mix | |||
| (NEBNext Quick Ligation Module) | New England Biolabs | E6056S | |
| (NEBNext Ultra II Ligation Module) | New England Biolabs | E7595S | |
| (Blunt/TA Ligase Master Mix) | New England Biolabs | M0367S | |
| Adapter mix | |||
| *Chemistry 9 | Oxford Nanopore Technologies | *Chemistry 9 EXP-AMII001 | |
| (Adapter Mix II [AMII]) | |||
| *Chemistry 14 | Oxford Nanopore Technologies | *Chemistry 14 EXP-NBA114 | |
| (Native adapter [NA]) | |||
| Sequencing | |||
| Flowcell priming kit | |||
| *Chemistry 9 | Oxford Nanopore Technologies | *Chemistry 9 EXP-FLP002 | |
| (Flush Buffer [FB]) | |||
| (Flush Tether [FT]) | |||
| *Chemistry 14 | Oxford Nanopore Technologies | *Chemistry 14 EXP-FLP004 | |
| (Flow Cell Flush [FCF]) | |||
| (Flow Cell Tether [FCT]) | |||
| Ligation Sequencing Kit | |||
| *Chemistry 9 | Oxford Nanopore Technologies | *Chemistry 9 SQK-LSK109 | |
| Adapter Mix (Adapter Mix [AMX]) | |||
| Ligation Buffer (Ligation buffer [LNB]) | |||
| Short Fragment Buffer (Short Fragment buffer [SFB]) | |||
| Sequencing Buffer (Sequencing Buffer [SQB]) | |||
| Elution Buffer (Elution buffer [EB]) | |||
| Loading Beads (Loading Beads [LB]) | |||
| Sequencing Tether (Sequencing Tether [SQT]) | |||
| *Chemistry 14 | Oxford Nanopore Technologies | *Chemistry 14 SQK-LSK114 | |
| Adapter Mix (Ligation Adapter [LA]) | |||
| Ligation Buffer (Ligation buffer [LNB]) | |||
| Short Fragment Buffer (Short Fragment buffer [SFB]) | |||
| Sequencing Buffer (Sequencing Buffer [SB]) | |||
| Elution Buffer (Elution buffer [EB]) | |||
| Loading Beads (Loading Beads [LIB]) | |||
| Sequencing Tether (Flow Cell Tether [FCT]) | |||
| Library solution (Library solution [LIS]) | |||
| Flush buffer (Flow Cell Flush [FCF]) | |||
| Flow Cell | |||
| *Chemistry 9 | Oxford Nanopore Technologies | *Chemistry 9 FLO-MIN106D | |
| (Flow Cell [R9.4.1]) | |||
| *Chemistry 14 | Oxford Nanopore Technologies | *Chemistry 14 FLO-MIN114 | |
| (Flow Cell [R10.4.1]) | |||
| Flow Cell wash | |||
| Flowcell wash kit (Flow cell wash kit) | Oxford Nanopore Technologies | EXP-WSH004 | |
| Consummables | |||
| Surface decontaminant | |||
| (DNA Away Surface Decontaminant, Squeeze Bottle [Molecular Bio]) | Thermofisher scientific/Fisher scientific | 7010PK | |
| (RNase Away Surface Decontaminant, Bottle [Molecular Bio]) | Thermofisher scientific/Fisher scientific | 7002PK | |
| PCR 8-Tube Strip 0.2ml, individual cap (PCR 8-Tube Strip 0.2ml, with Individual attached Flat Caps, Sterile, DNAse/RNAse, Pyrogen Free,Natural [Greiner]) | Greiner | 608281 | |
| PCR Tube 0.2ml (PCR Tube 0.2ml, Natural [Domed Cap] Bagged in 500s, Non-Sterile [Greiner]) | Greiner | 671201 | |
| 1000µL Filter Tips (500) (Stacked 1000µL Filter Tips [500]) | Thermofisher scientific/Fisher scientific | 11977724 | |
| 100µL Filter Tips (1000) | Thermofisher scientific/Fisher scientific | 11947724 | |
| 10µL Filter Tips (1000) (Stacked 100µL Filter Tips [1000]) | Thermofisher scientific/Fisher scientific | 11907724 | |
| Reinforced tubes tubes (2ml) with screw caps and o-rings (Fisherbrand™ Bulk tubes) | Thermofisher scientific/Fisher scientific | 15545809 | |
| Microcentrifuge tube (1.5ml) (1.5 ml Eppendorf Tubes [500]) | Eppendorf | 1229888 | |
| DNA LoBind Tubes (1.5ml) (DNA LoBind Tubes) | Thermofisher scientific/Fisher scientific | 10051232 | |
| Cryobabies labels | |||
| Gloves (S/M/L) | |||
| Paper towel |
References
- Rupprecht, C. E. Rhabdoviruses: rabies virus. Medical Microbiology. , University of Texas Medical Branch. Galveston, TX. (1996).
- Rabies. World Health Organization. , Available from: https://www.who.int/news-room/fact-sheets/detail/rabies (2023).
- Health Organization, W. orld WHO Expert Consultation on Rabies: WHO TRS N°1012. World Health Organization. , Available from: https://www.who.int/publications-detail-redirect/WHO-TRS-1012 (2018).
- Benavides, J. A., et al. Defining new pathways to manage the ongoing emergence of bat rabies in Latin America. Viruses. 12 (9), 1002(2020).
- Hampson, K. Estimating the global burden of endemic canine rabies. PLoS Neglected Tropical Diseases. 9 (4), e0003709(2015).
- Global genomic surveillance strategy for pathogens with pandemic and epidemic potential, 2022-2032. World Health Organization. , Available from: https://www.who.int/publications-detail-redirect/978924004679 (2022).
- Tsai, K. J., et al. Emergence of a sylvatic enzootic formosan ferret badger-associated rabies in Taiwan and the geographical separation of two phylogenetic groups of rabies viruses. Veterinary Microbiology. 182, 28-34 (2016).
- Chiou, H. -Y., et al. Molecular characterization of cryptically circulating rabies virus from ferret badgers, Taiwan. Emerging Infectious Diseases. 20 (5), 790-798 (2014).
- Sabeta, C. T., Mansfield, K. L., McElhinney, L. M., Fooks, A. R., Nel, L. H. Molecular epidemiology of rabies in bat-eared foxes (Otocyon megalotis) in South Africa. Virus Research. 129 (1), 1-10 (2007).
- Scott, T. P. Complete genome and molecular epidemiological data infer the maintenance of rabies among kudu (Tragelaphus strepsiceros) in Namibia. PLoS One. 8 (3), e58739(2013).
- Lembo, T., et al. Exploring reservoir dynamics: a case study of rabies in the Serengeti ecosystem. The Journal of Applied Ecology. 45 (4), 1246-1257 (2008).
- Coetzee, P., Nel, L. H. Emerging epidemic dog rabies in coastal South Africa: a molecular epidemiological analysis. Virus Research. 126 (1-2), 186-195 (2007).
- Ou de Munnink, B. B. First molecular analysis of rabies virus in Qatar and clinical cases imported into Qatar, a case report. International Journal of Infectious Diseases. 96, 323-326 (2020).
- Smith, J., et al. Case report: Rapid ante-mortem diagnosis of a human case of rabies imported into the UK from the Philippines. Journal of Medical Virology. 69, 150-155 (2003).
- Mahardika, G. N. K., et al. Phylogenetic analysis and victim contact tracing of rabies virus from humans and dogs in Bali, Indonesia. Epidemiology and Infection. 142 (6), 1146-1154 (2014).
- Tohma, K., et al. Molecular and mathematical modeling analyses of inter-island transmission of rabies into a previously rabies-free island in the Philippines. Infection, Genetics and Evolution. 38, 22-28 (2016).
- Tohma, K., et al. Phylogeographic analysis of rabies viruses in the Philippines. Infection, Genetics and Evolution. 23, 86-94 (2014).
- Saito, M., et al. Genetic diversity and geographic distribution of genetically distinct rabies viruses in the Philippines. PLoS Neglected Tropical Diseases. 7 (4), e2144(2013).
- Biek, R., Henderson, J. C., Waller, L. A., Rupprecht, C. E., Real, L. A. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceedings of the National Academy of Sciences. 104 (19), 7993-7998 (2007).
- Reddy, G. B., et al. Molecular characterization of Indian rabies virus isolates by partial sequencing of nucleoprotein (N) and phosphoprotein (P) genes. Virus Genes. 43, 13-17 (2011).
- David, D., Dveres, N., Yakobson, B. A., Davidson, I. Emergence of dog rabies in the northern region of Israel. Epidemiology and Infection. 137 (4), 544-548 (2009).
- Benjathummarak, S. Molecular genetic characterization of rabies virus glycoprotein gene sequences from rabid dogs in Bangkok and neighboring provinces in Thailand, 2013-2014. Archives of Virology. 161 (5), 1261-1271 (2016).
- Denduangboripant, J., et al. Transmission dynamics of rabies virus in Thailand: implications for disease control. BMC Infectious Diseases. 5, 52(2005).
- Talbi, C., et al. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathogens. 6 (10), e1001166(2010).
- Bourhy, H., et al. Revealing the micro-scale signature of endemic zoonotic disease transmission in an African urban setting. PLoS Pathogens. 12 (4), e1005525(2016).
- Zinsstag, J., et al. Vaccination of dogs in an African city interrupts rabies transmission and reduces human exposure. Science Translational Medicine. 9 (421), (2017).
- Yakovleva, A., et al. Tracking SARS-COV-2 variants using Nanopore sequencing in Ukraine in 2021. Scientific Reports. 12, 15749(2022).
- Mannsverk, S., et al. SARS-CoV-2 variants of concern and spike protein mutational dynamics in a Swedish cohort during 2021, studied by Nanopore sequencing. Virology Journal. 19, 164(2022).
- Soufi, M., et al. Fast and easy nanopore sequencing workflow for rapid genetic testing of familial Hypercholesterolemia. Frontiers in Genetics. 13, 836231(2022).
- Cabibbe, A. M. Application of targeted next-generation sequencing assay on a portable sequencing platform for culture-free detection of drug-resistant tuberculosis from clinical samples. Journal of Clinical Microbiology. 58 (10), 00632(2020).
- Xu, Y., et al. Nanopore metagenomic sequencing of influenza virus directly from respiratory samples: diagnosis, drug resistance and nosocomial transmission, United Kingdom, 2018/19 influenza season. Euro Surveillance. 26 (27), 2000004(2021).
- Stubbs, S. C. B., et al. Assessment of a multiplex PCR and Nanopore-based method for dengue virus sequencing in Indonesia. Virology Journal. 17, 24(2020).
- Croville, G., et al. Rapid whole-genome based typing and surveillance of avipoxviruses using nanopore sequencing. Journal of Virological Methods. 261, 34-39 (2018).
- Theuns, S., et al. Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies porcine kobuvirus as an important enteric virus. Scientific Reports. 8, 9830(2018).
- O'Donnell, V. K., et al. Rapid sequence-based characterization of African swine fever virus by use of the Oxford Nanopore MinION sequence sensing device and a companion analysis software tool. Journal of Clinical Microbiology. 58, 01104-01119 (2019).
- Brito, A. F. Global disparities in SARS-CoV-2 genomic surveillance. Nature Communications. 13, 7003(2022).
- ONT login/register. Oxford Nanopore Technology. , Available from: https://nanoporetech.com/login-register (2023).
- Software Downloads. Oxford Nanopore Technology. , Available from: https://community.nanoporetech.com/downloads (2023).
- GitHub. , Available from: https://github.com/ (2023).
- Brunker, K. Artic-rabv. , Available from: https://github.com/kirstyn/artic-rabv (2022).
- Campbell, K. MADDOG: Method for Assignment, Definition and Designation of Global Lineages. , Available from: https://github.com/KathrynCampbell/MADDOG (2022).
- Campbell, K. RABV-GLUE. Centre for Virus Research. , Available from: https://github.com/KathrynCampbell/MADDOG (2022).
- Itokawa, K., Sekizuka, T., Hashino, M., Tanaka, R., Kuroda, M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS ONE. 15 (9), e0239403(2020).
- Davis, M. W., Jorgensen, E. M. ApE, A plasmid editor: A freely available DNA manipulation and visualization program. Frontiers in Bioinformatics. 2, 818619(2022).
- Döring, M., Pfeifer, N. openPrimeR: Multiplex PCR primer design and analysis. , (2023).
- Quick, J. Multiplex PCR method for MiniON and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. 12 (6), 1261-1276 (2017).
- Laboratory Techniques in Rabies. World Health Organization. 1, Available from: https://apps.who.int/iris/bitstream/handle/10665/310836/9789241515153-eng.pdf (2018).
- Lembo, T. Partners for Rabies Prevention. The blueprint for rabies prevention and control: a novel operational toolkit for rabies elimination. PLoS Neglected Tropical Diseases. 6 (2), e1388(2012).
- Terrestrial Manual Online Access. World Organization for Animal Health. , Available from: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/ (2023).
- Mauti, S. Field postmortem rabies rapid immunochromatographic diagnostic test for resource-limited settings with further molecular applications. Journal of Visualized Experiments. (160), e60008(2020).
- Patrick, E., et al. Enhanced rabies surveillance using a direct rapid immunohistochemical test. Journal of Visualized Experiments. (146), e59416(2019).
- Lembo, T., et al. Evaluation of a direct, rapid immunohistochemical test for rabies diagnosis. Emerging Infectious Diseases. 12 (2), 310-313 (2006).
- Marston, D. A., et al. Pan-lyssavirus real time RT-PCR for rabies diagnosis. Journal of Visualized Experiments. (149), e59709(2019).
- Brunker, K. DNA quantification using the Qubit fluorometer. , Available from: https://www.protocols.io/view/dna-quantification-using-the-qubit-fluorometer-bc6vize6 (2020).
- Quick, J. DNA quantification using the Quantus fluorometer. , Available from: https://www.protocols.io/view/dna-quantification-using-the-quantus-fluorometer-7pzhmp6 (2020).
- Guppy protocol. Nanopore Community. , Available from: https://community.nanoporetech.com/protocols/Guppy-protocol/v/gpb_2003_v1_revaq_14dec2018 (2023).
- Flow Cell Wash Kit (EXP-WSH004). Nanopore Community. , Available from: https://community.nanoporetech.com/protocols/flow-cell-wash-kit-exp-wsh004/v/wfc_9120_v1_revh_08dec2020 (2023).
- Core Pipeline - arctic pipeline. , Available from: https://artic.readthedocs.io/en/latest/minion/ (2023).
- Samtools. , Available from: https://www.htslib.org (2023).
- Campbell, K., et al. Making genomic surveillance deliver: A lineage classification and nomenclature system to inform rabies elimination. PLoS Pathogens. 18 (5), e1010023(2022).
- Brunker, K., et al. Rapid in-country sequencing of whole virus genomes to inform rabies elimination programmes. Wellcome Open Research. 5, 3(2020).
- Bull, R. A., et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nature Communications. 11, 6272(2020).
- Okeke, I. N., Ihekweazu, C. The importance of molecular diagnostics for infectious diseases in low-resource settings. Nature Reviews. Microbiology. 19 (9), 547-548 (2021).
- Inzaule, S. C., Tessema, S. K., Kebede, Y., Ouma, A. E. O., Nkengasong, J. N. Genomic-informed pathogen surveillance in Africa: opportunities and challenges. The Lancet Infectious Diseases. 21 (9), 281-289 (2021).
- Kennedy, L. Integrating contact tracing and whole-genome sequencing to track the elimination of dog-mediated rabies: an observational and genomic study. eLife. , (2023).
- Pallerla, S. R. Diagnosis of pathogens causing bacterial meningitis using Nanopore sequencing in a resource-limited setting. Annals of Clinical Microbiology and Antimicrobials. 21, 39(2022).
- Quick, J. Real-time, portable genome sequencing for Ebola surveillance. Nature. 530 (7589), 228-232 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved