Method Article
A Bead Movement Based Computational Framework for 3-Dimensional Analysis of Biofilm Material Heterogeneity
In This Article
Summary
We describe a method for analyzing and quantifying the movement pattern of 1 µm carboxylate beads through heterogeneous bacterial biofilms. Comparison of the movement patterns can be used to quantitate differences in material properties of biofilms.
Abstract
Differences in the material properties of bacterial biofilms have been observed in biofilms of different bacterial species, within the same species under different growth conditions and after treatment with matrix modifying molecules. To better quantitate the material properties of 3D biofilms, an experimental and computational workflow was developed and applied to examine differences between Enterococcus faecalis, Salmonella enterica serotype Typhimurium and Escherichia coli biofilms as well as the role of the amyloid curli in confirming rigidity to Enterobacteriaceae biofilms. The spatio-temporal dynamics of 1 µm carboxylate beads in biofilms were tracked in 20 µm 3D biofilms over 20 minutes. The 4D image stacks were processed using the Mosaic plugin in ImageJ to produce 3D trajectory data of bead movement. This trajectory data was analyzed with a newly developed Bead Evaluator toolbox, where movement data, including trajectory lifespans, bead velocities, cell densities along trajectories, and bounding box information were computed and stored in csv-files. This paper presents the workflow from experimental setup and image recording to bead trajectory computation and analysis. The structure of curli-containing biofilms resulted in more stable bead interactions and less bead movement than in curli-mutant and Enterococcal biofilms. Bead movement did not appear strongly dependent on cell density when measuring the bead velocity and trajectory bounding box volume, supporting the hypothesis that other material properties of the biofilms control the bead dynamics. This technique is widely applicable to quantitating differences in biofilms of different matrix compositions as well as biofilms before and after matrix-modifying treatments.
Introduction
Bacterial biofilms are ubiquitous as part of human microbiota and continuously interact with molecules. These molecules range in size from 1 nm antibiotics and 1-3 mm bacteria to larger particles of fiber in the gastrointestinal tract. The composition of single- or multispecies biofilms affects the material properties and thus the movement pattern of particles through biofilms1,2,3,4,5. One example is bacterial amyloids, which have a conserved, fibrillar cross-beta sheet structure6. Amyloid curli is expressed by enteric bacteria such as Escherichia coli and Salmonella enterica serotype Typhimurium and genes have been detected in multiple other bacterial phylum7. Various material properties of biofilms are affected by curli8,9. Curli directly interacts with other components of the matrix such as extracellular DNA (eDNA) and cellulose10,11. Curli surrounds the cells and affects cellular membrane rigidity12 and the overall viscoelastic properties of the biofilm13. Curli mediates increased tensile strength by binding to fibronectin, resulting in an increase in strong glass-surface attachment14. Incoming bacteriophages bind to curli and limit phage invasion into biofilms15.
When using multitest coated well slides to analyze roughly 20 µm thick Enterococcus faecalis, E. coli, and S. Typhimurium biofilms using confocal microscopy, clear differences between E. coli, S. Typhimurium10,16 and E. faecalis biofilms (current study) could be observed. While Enterobacteriaceae species biofilms had a high level of rigidity and areas with low cellular density were easy to image, obtaining clear high-resolution pictures of E. faecalis biofilms using line and frame averaging required the application of pressure to the slide to induce sufficient surface tension for cell stability during the imaging process. Bacterial amyloids such as curli form highly ordered structures, suggesting they may be relatively rigid17. This motivated the hypothesis that amyloid curli could be inducing rigidity in E. coli and S. Typhimurium biofilms. There was no clear evidence that E. faecalis was expressing amyloids under the conditions studied. The protein Esp, a pilin gene associated with more pathogenic strains of E. faecalis, was recently shown to produce amyloid structures18; however, using blastn and blastp searches, this gene was not detected in the E. faecalis commensal type strain OG1RF used in these studies. The pheromone cOB1, produced by OG1RF, can form amyloid-like structures19. However, with the given biofilm growth conditions and amyloid detection methods previously used for S. Typhimurium amyloid staining10 in E. faecalis, OG1RF amyloids could not be detected (data not shown). A new four-dimensional (4D) image technique was developed to compare the overall material properties amongst the viscous E. faecalis, E. coli and S. Typhimurium as well as to determine the contribution of amyloid to Enterobacteriaceae biofilms using amyloid mutants of S. Typhimurium and E. coli.
In the past, fluorescent beads were successfully used to analyze the material properties of biofilms in two dimensions (2D) using microrheology20,21,22,23,24,25. This can be applied to a three-dimensional biofilm by studying 2D optic slices at various depths in the biofilm26. The current technique was developed to track 1 µm microscale beads in 3D over time for use in 4D modeling. Part of the rationale was the overarching concept of using 4D modeling to understand movement of plasmids through gastrointestinal microbiota communities. Fluorescently charged carboxylate beads with a 1 µm diameter were used since these correspond well, with respect to size and charge, to E. faecalis, the chosen model organism for plasmid movement and maintenance27, 28. A 4D assay to quantify the physical properties of biofilms was developed (Figure 1A). In the devised methodology, beads were added to biofilms and their spatio-temporal trajectories were recorded through 10-20 µm thick biofilms over the course of 10-20 minutes. Bead trajectories in 3D were then quantified in terms of trajectory length, bead velocity, trajectory bounding box volume (minimal box containing the trajectory), and bounding box cellular density using a newly developed toolbox. The following protocol can be employed to generate 4D image data of bacteria and bead containing biofilms, to preprocess the data with ImageJ29 and the plugin Mosaic, and to analyze bead trajectories with a Bead Evaluator toolbox.
This technique has multiple applications for examining material properties as well as tracking particle and bacterial movement in three dimensions. For example, an early version of this technique was used to characterize the effect of monoclonal antibodies directed against curli on the structural integrity of biofilms16. The full version has multiple tools to provide a more detailed analysis of the biofilm material properties and is continuing to be used to examine effects of monoclonal antibody treatment on biofilms. Particles of different charges can be used to examine the material charge properties of the biofilms and movement of particles through biofilms with different matrix compositions. This could be used to compare the results from 2D microrheology that reveal the material properties responsible for movement of beads we observed in biofilms that were not under flow. This technique could also be used on mixed species biofilms with regions of different biofilm composition. Biofilms can be live imaged under flow in microfluidic devices and flow cells to examine changes in the material properties between static and flow biofilms as well as the effect of flow on movement of particles. The techniques can also be applied to fluorescently labeled bacteria to characterize the movement of exogenous bacteria through a biofilm community. Using three colors, fluorescently labeled donor bacteria, fluorescently labelled recipient bacteria and fluorescently labeled plasmids could be used to track movement, docking, and transfer of plasmids.
Protocol
1. Biofilm preparation
NOTE: Biofilms for analysis can be grown using any method that allows the biofilm to form on an optic glass surface. The biofilm structure should adhere to the optic surface enough that the structure is not disrupted during washing and/or mounting steps of the protocol. Below describes the technique for 96-well optic bottom plates and 12 mm glass coverslips in 24-well plates. Other options include different sized optic bottom plates and optic flow chambers with and without flow.
- Biofilm setup
- Add bacterial growth medium to the wells of the plate. For this study, for E. faecalis, add 2 mL of Todd-Hewitt (TH) to a 24-well plate and add 0.4 mL of TH to optic bottom 96-well plates. For S. Typhimurium, E. coli and the isogenic csgAB curli mutants, add 0.7 mL of no-salt Luria Broth (LB) to the wells. If using optic bottom chambers proceed to Step 1.2.
- Place 12 mm #1.5 glass optic coverslips in a Petri dish and cover them with ethanol.
- Using forceps, remove the coverslip and use a flame to light the remaining alcohol on fire. Allow the alcohol to burn off. Just use the flame to light them on fire; do not hold the coverslip in the flame because it will crack. Allow the coverslip cool for 10-20 s before placing it in the well to prevent cracking.
- Place the coverslip into the well containing the medium at an angle to prevent it from laying on top of the medium. Do not add the coverslip to a dry well and then add medium because this will cause the coverslip to stick to the bottom of the well.
- Use a sterile pipette tip to carefully push the coverslip to the bottom of the well containing the medium.
NOTE: Remember to match the thickness (#1 or #1.5) of the optic glass coverslip or optic bottom plate to the thickness of the confocal microscope optics.
- Incubate the biofilm under proper conditions for biofilm growth. In these studies, grow E. faecalis biofilms as static cultures at 37 °C and grow E. coli biofilms aerobically at 30 °C.
- In these studies, grow E. faecalis biofilms for 2 days with medium switches in the morning and early evening. To prevent damaging or dislodging the biofilm, carefully slant the plate. Place the pipette tip near the bottom edge of the well and slowly draw out the medium. Add the first mL of fresh medium in the same fashion. Add the second mL slowly near the medium/well interface.
- In these studies, use growth conditions for optimal curli production. Grow S. Typhimurium biofilms at 28 °C for 6-8 days incubated on a slant that allowed the biofilm to attach around 2/3 of the way up the slide and then grow as a pellicle on the air-liquid interface. This was done without a medium change. To prevent the medium from drying out, place the 24-well plate in a chamber with a water pan.
2. 4D imaging
- Preparation of the biofilm mount
- Dilute Crimson 1 µm carboxylate FluoSpheres beads 1:50 in PBS (2 x 107 beads in 1 mL of PBS). If using optic bottom chambers, add Syto9 at a dilution of 1 µL to 300 µL of bead preparation.
- (optional) Wash the biofilm to remove traces of growth medium if the growth medium has autofluorescence. In these experiments, wash twice with 1 mL of PBS, carefully slanting the plate, placing the pipette tip near the bottom edge of the well and slowly drawing out the medium. Add PBS by placing the tip near the bottom edge and slowly fill the well. Use this technique on optic bottom wells in addition to coverslips in a 24-well plate.
- Remove medium or PBS. Add diluted Crimson beads prepared in 2.1.1 to the biofilm. In these studies, add 1 mL of beads (2x107) to the coverslips and 0.2 mL of beads (4x106) and Syto9 to 96-well optic bottom plates.
- Incubate for 1 min at room temperature to allow bead association.
- Remove beads and gently wash the biofilm once with PBS to remove unassociated beads. In these studies, for the coverslip, gently wash with 1 mL of PBS, refill the well with 1 mL PBS, and proceed to 2.1.6. For the 96-well optic bottom plates, gently wash the biofilm with 0.2 mL of PBS and refill the well with 0.2 mL of PBS. The optic bottom chamber is now ready to image, so proceed to step 2.2.
- (If using a coverslip) Add 1 µL of Syto9 (green fluorescent DNA stain; diluted per manufacturer's instructions) into the center of a well on a coated 10-well multitest slide. These coated coverslips have well depths of 23-25 µm.
- (If using a coverslip) Using alcohol flame sterilized forceps, carefully remove the coverslip from the well and invert onto the well containing the Syto9 drop. Leaving the coverslip in 1 mL of PBS makes it easier to remove from the well and helps wash unassociated beads out of the biofilm.
- (If using a coverslip) Carefully seal the coverslip with finger-nail polish without sliding or pressing down on the coverslip, which can cause surface tension that will stop movement in more viscous, less rigid, biofilms.
- (If using a coverslip) Allow the nail polish to dry. Carefully wipe the outer surface of the coverslip with 70% ethanol. Wipe without apply any pressure to the coverslip for reasons stated above.
- Confocal imaging
NOTE: In these studies, use an inverted spectral imaging laser scanning confocal microscope equipped with a TCS confocal system with the 63x objective. The scope will be used to generate a 4D video (Figure 1). The 3D biofilm will be composed of Z slices captured in 0.5 µm steps through biofilms which are 18-20 µm thick, generating 36-40 Z slices. Each 3D biofilm will take 50-60 s to capture. The slices together will make up a frame, which can be visualized as a 3D biofilm. This process will be repeated 20 times to generate the 4D time lapse video for a total tracking time of 18-20 minutes.- Set the scope to capture fluorophores. In these studies, excite Syto9 (bacterial DNA staining) with a 488 nm laser and measure emission from 495 to 540 nm (the Leica Sp5 is a spectral imaging microscope). Excite crimson (red) beads with a 633 nm laser and measure emission from 650 nm to 700 nm. These fluorophore settings can be adjusted to capture any desired fluorophores.
- Choose the xyzt imaging mode.
- Identify a region of the biofilm with a mixture of higher and lower density regions to capture differences in viscoelastic properties in thick and thin regions of the biofilm.
- Set a Z-stack of 18-20 µm thick. In a coverslip mounted biofilm, avoid the very top and very bottom of the biofilm touching against the glass to avoid bead trapping artifacts (please see discussion).
- Adjust the gain and offset to use the entire dynamic range of the intensity in the brightest spot of the biofilm. This minimizes signal overlap from lower layers of the biofilm.
- Set the slice thickness to 0.5 µm. This allows rapid imaging without the loss of bead information.
- Set resolution to 512 x 512 (0.48 µm). This allows rapid imaging yet generates images with sufficient resolution to see the biofilm structure and bead movement details.
- Minimize imaging time.
- Set to capture 20 stacks.
- Save as .lif (or similar confocal file). The 4D movie can be generated in ImageJ.
3. Generating the 4D biofilm video with ImageJ
- Open the .lif file in ImageJ with the following settings: View stack with Hyperstack, Stack order XYCZT, color mode: colorized, autoscale checked. Then press OK.
- Select Image > Color > Split Channels.
- Select Image > Color > Merge Channels > Create Composite > OK.
- Select Plugins > 3D viewer > Channels uncheck blue > OK.
- Select Edit > Show content checked > Show bounding box checked.
- Use the mouse click and hold to select the image. Then rotate the image while continuing to hold down on the click button. Rotate the image so the bottom of the biofilm is on the bottom and the angle supports visualizing the beads. Then release the image.
- Press the red Record button at the bottom of the window to record a video.
- Save as .avi-file using jpeg as the compression.
4. Generating trajectory data
- Install the open-source tool ImageJ (https://imagej.net/Fiji) and the particle tracking plugin Mosaic (https://imagej.net/MOSAICsuite).
- Import the .lif file containing the two channels for beads and bacteria. Split the channels and save the files separately.
- In ImageJ, store the image voxel size (x, y, and z dimensions) and the time step size to a text file.
- In ImageJ, go to Plugins | Mosaic and launch the Particle Tracker 2D/3D.
- Input the following parameters: Radius: 3, Cutoff: 0.003, Per/Abs: 0.12, Link Range: 2, Displacement: 10.00 with the dynamics set at Brownian. Please see results for rationale behind the choice of Brownian in these experiments.
- Generate trajectories and export the trajectory list as .csv files.
5. Analyzing trajectories
- Install VRL Studio (https://vrl-studio.mihosoft.eu)
- Download Biofilm project (https:// neurobox3d.github.io/Biofilm/) and launch in VRL Studio
- Load the trajectory file in ImportData
- Specify the x, y, and z pixel size (use ImageJ to locate these values) in ProcessTrajectories.
- Specify the frame interval in ComputeVelocity (use ImageJ to locate this value).
- Load the bacterial tiff-file (see step 3.2) in Comdensity.
- Set output paths for velocity data in SaveVelocityDataToFile and trajectory data in SaveTrajectoryDataToFile.
- Invoke SaveVelocityDataToFile and SaveTrajectoryDataToFile.
- Import data to Excel for analysis. These data will include trajectory lengths, trajectory lifespans, trajectory bounding box dimensions and volumes, average bead velocities and variances. The analysis computes weighted variables using the channel for Syto9 labelled bacteria to compute the local (within given trajectory bounding boxes) cellular densities. The analysis results in calculation of weighted average velocities and variances as well as weighted averages and variances of bounding box variance.
Results
This method was used to test the hypothesis that curli may confer rigidity to E. coli and S. Typhimurium biofilms, reducing bead movement during confocal microscopy experiments. The current toolbox was used to compare the material properties of Enterococcus faecalis commensal type strain OG1RF to Salmonella enterica serotype Typhimurium, E. coli, and their respective isogenic curli mutants (Figure 1B and Supplemental Video 1, Supplemental Video 2, Supplemental Video 3, Supplemental Video 4, Supplemental Video 5, Supplemental Video 6). Biofilm material properties could potentially differ with respect to rigidity (e.g., curli bound to eDNA) or electrostatic and hydrophobic interactions between the negatively charged beads and the biofilm cells and matrix materials as well as cellular density.
Reproducibility
The Biofilm toolbox was programmed in Groovy30 and Java31 within VRL-Studio32 enabling a modular workflow design with automatic User Interface (UI) generation of all computational components. This allowed for an automated workflow, removing unintentional experimenter-induced bias when analyzing the results.
Use of MSD to confirm type of motion in the biofilms
For analysis of trajectories using Particle Tracker 2D/3D, different dynamics settings for analysis of different bead movement types are available. For these studies, the setting "Brownian motion" (i.e., diffusion-driven movement) was chosen since E. faecalis is a non-motile bacterium, E. coli and Salmonella do not express flagella in biofilms, and the experiments were performed in a closed system in the absence of flow. This setting could be further validated by the computed mean square displacements (MSD) of the beads. Using the definition 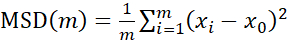 where m is the number of trajectory segments, the change of the MSD over the course of each trajectory can be computed. Linear trajectories indicate diffusive bead movement (Figure 2A). Using quadratic least squares fitting, the average movement pattern of all the beads in the biofilm was computed, showing dominant linear order and validating passive diffusion as the driving force (Figure 2A-2F).
where m is the number of trajectory segments, the change of the MSD over the course of each trajectory can be computed. Linear trajectories indicate diffusive bead movement (Figure 2A). Using quadratic least squares fitting, the average movement pattern of all the beads in the biofilm was computed, showing dominant linear order and validating passive diffusion as the driving force (Figure 2A-2F).
Bounding box analysis.
The toolbox uses ImageJ Mosaic and Particle Tracker 2D/3D to generate trajectories (Step 4) and then using the automated Biofilm analysis pipeline, generates important data about the bead trajectories that can be used to compare biofilm material properties. The bounding box volume in µm3 was measured by constructing the minimal box that contains a trajectory and measuring its volume (Figure 3).
E. faecalis biofilms have more bead movement with bounding box values of 1-6000 µm3 (Figure 3B, 3C and 3D). The results confirm that the movement seen in a glass coverslip mounted onto a coated slide with an approximately 25 µm coated well (Figure 3C) versus biofilms grown in the bottom of optic glass wells and directly imaged (Figure 3D) yield equivalent results with few differences. The only difference was that near the top of the mounted coverslip of E. faecalis biofilms stable trajectories with lifespans greater than 10 minutes but at the same time small bounding boxes could be registered, whereas in the optic bottom plate a select number of beads with higher mobility could be registered. Taken together, this suggests that mounting the glass slide may have changed the system's surface tension at the top of the biofilm against the slide in the mounted coverslip biofilm, which ultimately decreased mobility of some beads in less viscous biofilm regions (Figure 3B, 3C, 3D and 3I). The trajectories that fall into this category happened to be a very small percentage and, even with this small number of trapped beads, the average MSD of the E. faecalis on a mounted coverslip was slightly higher than the MSD calculated from an optic bottom plate biofilm (Figure 3).
S. Typhimurium and E. coli bead trajectories had smaller bounding box volumes of 0-10 µm3 (Figure 3A, 3B, 3E and 3F), compared to isogenic curli mutants with bounding boxes of 1-6000 µm3 for E. coli and 1-5000 µm3 for S. Typhimurium (Figure 3A, 3B, 3F and 3H), demonstrating larger bead mobility. These results suggested that the presence of the amyloid correlated with increased rigidity in the biofilms and was consistent with the lack of notable biofilm movement in the videos. The bounding box volumes were consistently small (0-10 µm3) even in low density regions of the biofilm. This observation is consistent with previous observations that curli can be present in low cell density regions of the biofilm10.
It was not possible to compare the behavior of Enterobacteriaceae biofilms on optic bottom plates because they grow as pellicles on the air-liquid interface (Step 1.2.2). When using a coverslip, the pellicle attached to the coverslip at the interface and when the coverslip was removed the pellicle laid on the coverslip creating a single image surface. In an optic bottom plate grown at a slant, imaging was done with liquid still in the well. This means the pellicle is still floating above the optic bottom and makes the pellicle out of the working depth of an inverted scope such as the Leica Sp5. Removing enough medium to bring the biofilm into the working depth of the microscope caused the specimen to dry out over the 20-minute imaging process.
Overall, the plots confirm the visual observations in the supplemental movies and are consistent with the MSD differences observed (Figure 3I and 3J).
Trajectory lifespans
The trajectory lifespan was measured as the number of consecutive frames in which a bead was registered (Figure 3).
In the more viscous, fluid-like E. faecalis biofilms, all beads had a trajectory lifespan of shorter than 10 minutes and the majority of trajectories ranged between 2-5 minutes for E. faecalis biofilms. However, beads with registered short trajectory lifespans could be located by visual inspection in E. faecalis biofilms over the total imaging time-window (Supplemental Video 1 and 2). Thus, it is possible that beads move along a registered trajectory, intermittently disassociating from the biofilm and terminating a trajectory, and reassociating with the biofilm, at which point a new trajectory is initiated. This ultimately would lead to short trajectory lifespans under continuous presence of beads in the biofilm. It is important to note that using this technique, the trajectory lifespans, especially in a viscous biofilm, tends to underestimate the total time a bead is associated with the biofilm.
In S. Typhimurium biofilms, which had smaller bounding box volumes, a majority of beads (around 80%) had long trajectory lifespans of 16-20 frames, corresponding to roughly 15-20 minutes real-time (Figure 3A, 3G and 3H). Contrary to these, isogenic curli mutant biofilms carried more mobile beads with bounding box volumes ranging between 1-6000 µm3 (E. coli) and 1-5000 µm3 (S. Typhimurium) (Figure 3A, 3B, 3F and 3H). In contrast to E. faecalis biofilms with >70% trajectories having bounding box volumes larger than 10 µm3, however, Enterobacteriaceae species biofilms registered only 30% bead trajectories with bounding box volumes above 10 µm3. Even though overall bead trajectory lifespans were smaller in curli mutant biofilms, some trajectories reflected substantial bead movement and long trajectory lifespans (Figure 3H). This observation could indicate that this variability may correspond to varying biofilm material properties, such as viscoelasticity, and/or particle surface chemistry changes such as charge.
Analysis of bead trajectory lengths and velocities
The trajectory length is a measurement of the distance traveled by beads in µm. This measurement is consistent with the velocity of bead movement in µm/s. Consistent with the larger bounding box volumes, beads in E. faecalis biofilms had 10-fold longer trajectories, 5-20 µm, versus <4 µm in biofilms containing curli. Consistent with the shorter trajectories (Figure 4A). E. faecalis beads measured up to 15x higher velocities with a majority of beads having velocities in the range of 0.01-0.15 µm/s versus velocities <0.006 µm/s (Figure 4B). Nonetheless, curli mutant biofilms measured overall lower velocities and shorter trajectories compared to E. faecalis biofilms, but longer trajectories and higher velocities than the curli containing parental strains (Figure 4A and 4B).
Noteworthy is the fact that the fibrillar lattice-like structure of curli30 may affect mobility in an anisotropic way, reducing movement in the xy-plane and allowing increased mobility in the z-direction (Figure 2G). The large trajectory pool (approximately 800) belonging to roughly 50 unique beads in curli-containing biofilms would be consistent with the limitations of Mosaic Particle Tracking, counting each one of these rapidly moving beads as a single bead in x, y and z. Additional research and software development will be necessary to confirm this observation.
Analysis of bead movement dependence on cellular density
The dependence of bead movement on cellular density was determined by using weighted average velocities and variances, as well as averages/weighted averages and variances of bounding box volumes. The second imaging channel for Syto9 labelled bacteria was used to compute the local cellular densities in the calculation of weighted velocities. The cellular density was computed by averaging the Syto9 voxel data over the bounding box of each trajectory edge (Figure 5, right). Thus, the bead velocity can be weighted by the edge-wise (local) cell densities. There are multiple types of stains that could be used to visualize bacteria including stains for the cell wall, membranes, and DNA content. To determine cellular density Syto9 was chosen because it gives the most consistent signal no matter which optical Z slice is being visualized. Envelope stains (cell wall and membrane) will give a different signal depending on the position of the Z slice. If the Z slice includes the top or bottom of the cell, the signal will be stronger than if the Z slice is through the middle of the cell where only the outline of the cell is stained.
The bead trajectories from the red channel were tracked for 20 frames, where individual trajectories had a minimal lifespan of 2 frames and a maximum of 20 frames with 19 trajectory segments connecting the frames (Figure 5). To study cell density dependence of bead mobility, the GFP intensity per voxel (each of the individual measurements in the 512x512 voxel image) was determined. The cell density around each segment of the bead trajectory was calculated as the locally averaged density in the bounding box of the segment.
For some biofilms, statistically significant density-dependence could be documented (Figure 6), most prominently for E. faecalis biofilms that were grown on a glass coverslip and inverted onto a multiwell slide (Figure 6A). To the contrary, E. faecalis biofilms that were grown on the bottom of a 96 well plate (Figure 6B) showed no density dependence. In conclusion, this suggests that highly fluid E. faecalis biofilms could potentially be slightly compressed due to the mounting on a multi-well slide, which is consistent with a reduction in the number of beads moving more rapidly and those with small bounding box volumes that are trapped at the top of the biofilm against the glass slide (Figure 3C vs Figure 3D). Both the Salmonella and E. coli biofilms (Figure 6C and 6D) and their isogenic mutants (Figure 6E and 6F) showed minor to no cellular density dependence.
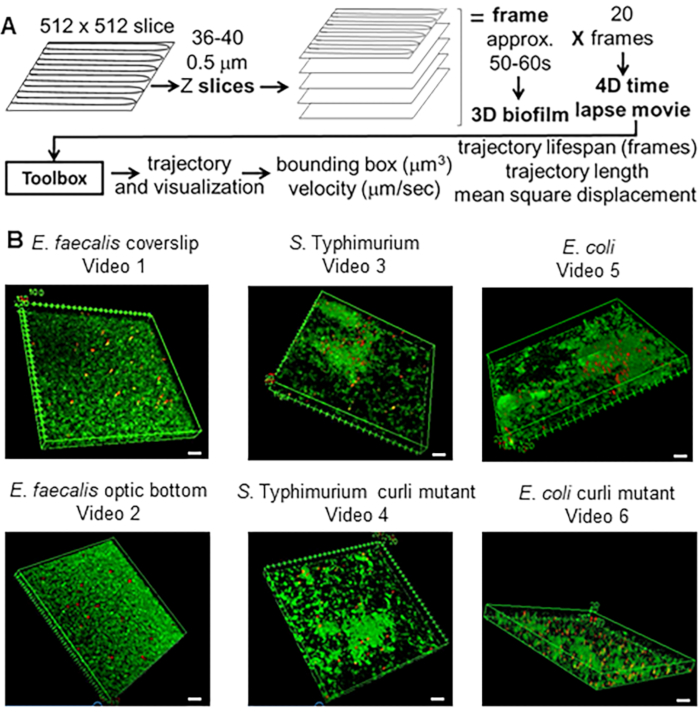
Figure 1. Imaging and analysis pipeline (steps 2-4) (A) Biofilms are imaged as outlined in 2.2. Using the imaged biofilms (see 3) bead trajectories were generated as described in 4. Using the trajectories relevant data was computed with the analysis toolbox (see 5) (B) Biofilms were grown as described in step 1 on coverslips (E. faecalis, Video 1), S. Typhimurium (Video 3), E. coli (Video 5) and isogenic curli mutants (Video 4, Video 6) or in an optic bottom 96-well plate (E. faecalis bottom, Video 2). The white scale bar is 20 mm. This figure is reproduced with permission from (31). Please click here to view a larger version of this figure.
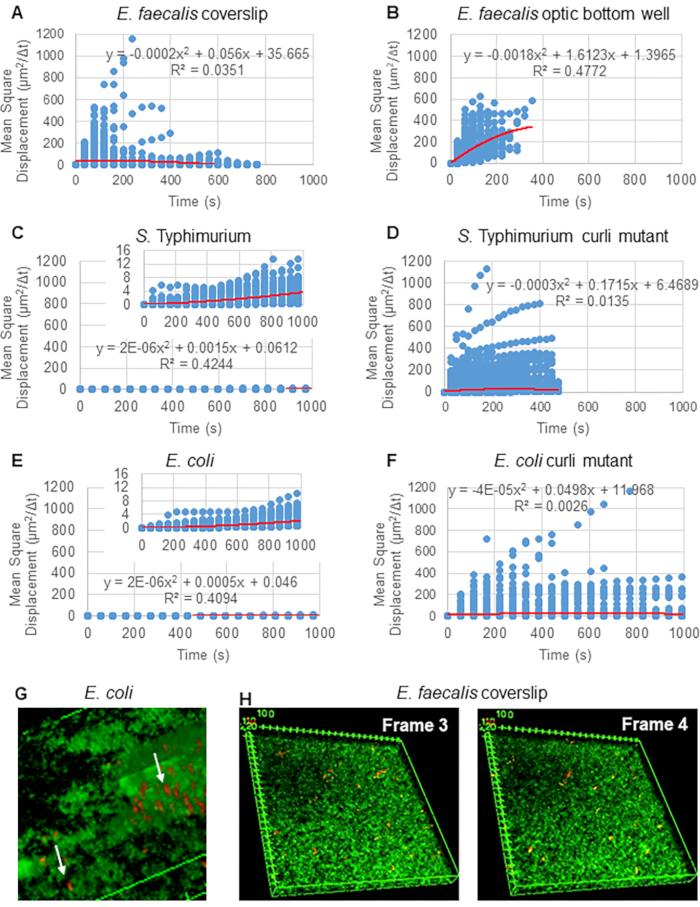
Figure 2. The bead movement in the biofilms were determined to be diffusive in nature (Brownian motion, see step 5) (A-F) MSD data shows linear behavior (red line) to validate Brownian motion. Biofilms were grown according to step 1 (G) Example of elliptical bead movement observed in E. coli and S. Typhimurium biofilms taken from one frame of the E. coli 4D biofilm assay (H) Example of large changes in bead patterns between frame 3 and 4 taken from E. faecalis optic bottom well. Note the biofilm itself elicits some flow (Video 1 and 2), which makes it appear that frame 3 and 4 are differently oriented. This figure is reproduced with permission from (31). Please click here to view a larger version of this figure.
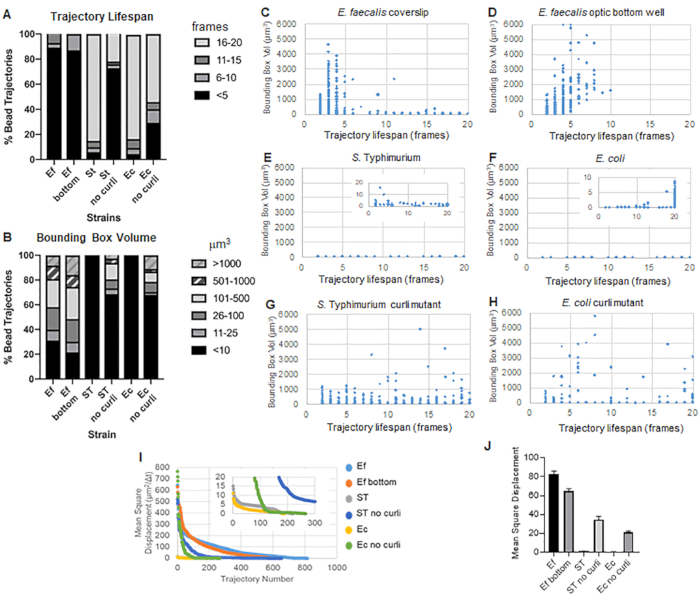
Figure 3. Analysis of differences in rigidity using bounding boxes and trajectory lifespans. Biofilms were grown as described in step 1 on coverslips (E. faecalis, Video 1), S. Typhimurium (Video 3), E. coli (Video 5) and isogenic curli mutants (Video 4, Video 6) or in an optic bottom 96-well plate (E. faecalis bottom, Video 2). Trajectory lifespans are presented in % of total bead trajectories (A) and scatter graphs (C-H), along with bounding box volumes (computed in step 5) (I) Comparison of the bead MSDs in the different biofilms (H) Average MSDs of each type of biofilm. The bars indicate the 95% confidence interval of the data. This figure is reproduced with permission from (31). Please click here to view a larger version of this figure.
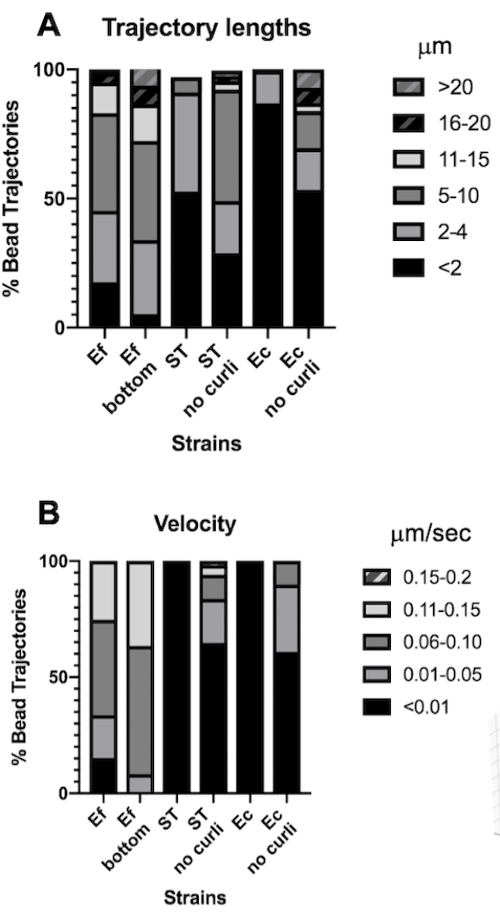
Figure 4. Analysis of differences in trajectory length and bead velocity. Biofilms were grown as described in step 1. Trajectory lengths are shown in µm and are presented as % of total bead trajectories (A). Velocity is shown in µm/s and presented as % of total bead trajectories (B). This figure is adapted with permission from (31). Please click here to view a larger version of this figure.
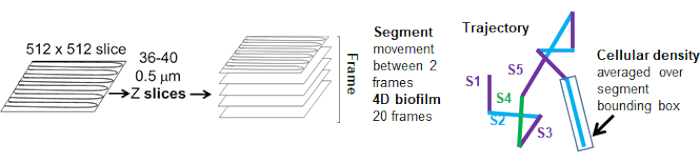
Figure 5. Outline of trajectory segment analysis. This figure is adapted with permission from (31). Please click here to view a larger version of this figure.
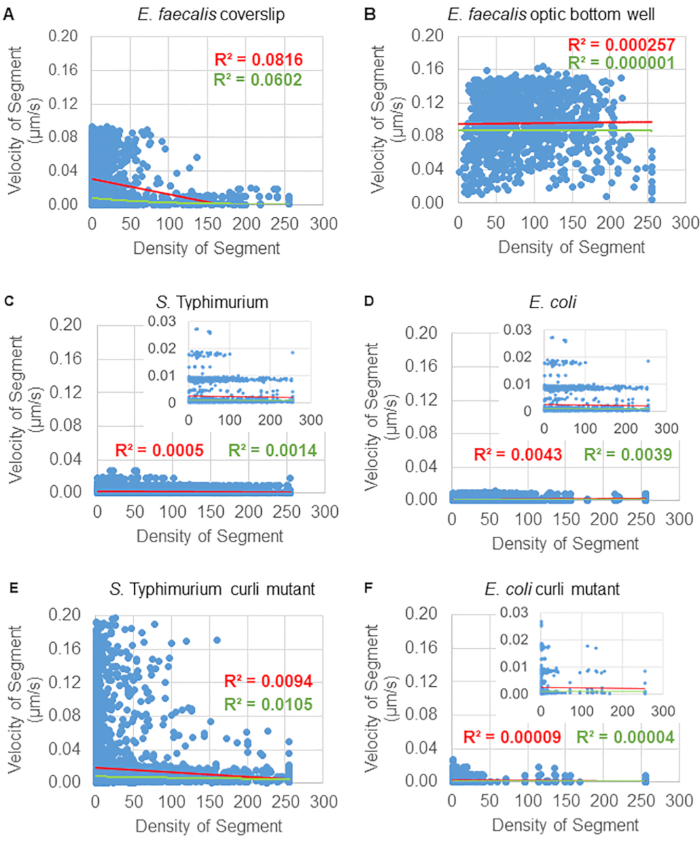
Figure 6. Studying the effect of biofilm density on bead velocity using the biofilm analysis pipeline (step 5). Results indicated that bead velocity is not exclusively cell density dependent. Biofilms were grown as described in step 1. Trajectories were analyzed at the scale of individual trajectory segments (Figure 5). For each segment, bead velocity in µm/s was plotted against bounding box cellular density (average GFP per voxel within the bounding box). The red line shows the linear regression, and the green line the exponential regression trace. This figure is reproduced with permission from (31). Please click here to view a larger version of this figure.
Supplemental Video 1. 4D video of 24-hour E. faecalis OG1RF biofilm grown on a 1.5 thick optic glass coverslip. The 4D time lapse video was generated using a microscope at 512x512 resolution. A Z-series of 40 images was generated by imaging an approximately 20 µm thick region of a biofilm in 0.5 µm steps. Each Z series was one frame and required 50-60 s to capture. A series of 20 contiguous frames were captured to produce a 4D video. The video playback is at approximately 120x. Video is representative of at least 6 independent experiments. Please click here to download this video.
Supplemental Video 2. 4D video of a 24-hour E. faecalis OG1RF biofilm grown on a 96-well optic bottom plate. The 4D time lapse video was generated using a microscope at 512x512 resolution. A Z-series of 40 images was generated by imaging an approximately 20 µm thick region of a biofilm in 0.5 µm steps. Each Z series was one frame and required 50-60 s to capture. A series of 20 contiguous frames were captured to produce a 4D video. The video playback is at approximately 120x. Video is representative of 3 independent experiments. Please click here to download this video.
Supplemental Video 3. 4D video of a Salmonella enterica serotype Typhimurium biofilm ATCC 14028 grown on 1.5 thick optic glass coverslips for 6-7 days. The 4D time lapse video was generated using a microscope at 512x512 resolution. A Z-series of 40 images was generated by imaging an approximately 20 µm thick region of a biofilm in 0.5 µm steps. Each Z series was one frame and required 50-60 s to capture. A series of 20 contiguous frames were captured to produce a 4D video. The video playback is at approximately 120x. Video is representative of 3 independent experiments. Please click here to download this video.
Supplemental Video 4. 4D video of Salmonella enterica serotype Typhimurium biofilms ATCC 14028 curli (csgBA) mutant were grown on 1.5 thick optic glass coverslips for 6-7 days. The 4D time lapse video was generated using a microscope at 512x512 resolution. A Z-series of 40 images was generated by imaging an approximately 20 µm thick region of a biofilm in 0.5 µm steps. Each Z series was one frame and required 50-60 s to capture. A series of 20 contiguous frames were captured to produce a 4D video. The video playback is at approximately 120x. Video is representative of 3 independent experiments. Please click here to download this video.
Supplemental Video 5. 4D video of E. coli UTI89 grown on 1.5 thick optic glass coverslips for 6-7 days. The 4D time lapse video was generated using a microscope at 512x512 resolution. A Z-series of 40 images was generated by imaging an approximately 20 µm thick region of a biofilm in 0.5 µm steps. Each Z series was one frame and required 50-60 s to capture. A series of 20 contiguous frames were captured to produce a 4D video. The video playback is at approximately 120x. Video is representative of 3 independent experiments. Please click here to download this video.
Supplemental Video 6. 4D video E. coli UTI89 curli (csgBA) mutant grown on 1.5 thick optic glass coverslips for 6-7 days. The 4D time lapse video was generated using a microscope at 512x512 resolution. A Z-series of 40 images was generated by imaging an approximately 20 µm thick region of a biofilm in 0.5 µm steps. Each Z series was one frame and required 50-60 s to capture. A series of 20 contiguous frames were captured to produce a 4D video. The video playback is at approximately 120x. Video is representative of 3 independent experiments. Please click here to download this video.
Discussion
Critical steps and troubleshooting
The biggest challenge of this technique is using a mounted coverslip with a very viscous biofilm like E. faecalis. The coverslip needs to be carefully and accurately placed on the multiwell slide without repositioning it. During the sealing step, care needs to be used to prevent pushing down on the coverslip or accidently pushing/sliding it across the slide surface. Any movement or pressure may create surface tension and block movement of a viscous biofilm. If possible, comparing biofilm material properties by imaging a biofilm on an optic bottom well to a coverslip mount will allow technique assessment. When correctly performed, a coverslip mount very closely resembled a biofilm in an optic bottom plate for E. faecalis.
In addition, when using a mounted coverslip, imaging of the interfaces of the biofilm with the coverslip at the bottom or the slide at the very top should be avoided. When using an inverted scope, with the coverslip at the bottom, there can be trapped beads at the base of the biofilm against the coverslip. These beads pass through the biofilm and become trapped against the coverslip even after gentle washing. They have x, y and z coordinates of 0 and bounding box coordinates of 0. However, for certain applications, such as examining biofilm integrity after treatment, these data points can be used as a tool. The ability of beads to penetrate through a thick region of biofilm to the bottom of coverslip can be used to assess biofilm integrity after treatment (manuscript in preparation in collaboration with Tükel laboratory). At the top of the biofilm, in a viscous biofilm like E. faecalis we had some evidence of compaction imposed by the coverslip. This limited the movement of some beads at the glass slide interface and may have introduced some density dependence to the bead movement analysis.
The washing steps were necessary for the biofilms because the growth medium has strong autofluorescence in the green channel. We choose to use excess beads and remove unassociated beads by washing to maximize the associated beads to get the most accurate characterization of the observed regions.
The number of beads and washes needed to obtain desired data sets needs to be determined empirically. The presence of too many beads in a biofilm generates impossibly large data sets that are hard to analyze. The presence of too few beads does not generate a thorough sampling of the biofilm environments. However, control of the number of beads added (2x107 beads in 1 mL of PBS) and the use of wash steps, resulted in a relatively consistent number of beads (40-140) associating with the biofilm depending on its structure, spatial arrangement, and composition.
When studying biofilms with mixed viscous and rigid regions, beads can become trapped in the rigid regions over time. In this case, imaging needs to be started immediately after addition of the beads. This often cannot be accomplished using coverslips but requires optic bottom plates or flow cells where imaging can be done immediately after addition of the beads and the wash step(s).
Modifications and Future Applications
Use of microfluidic devices. In our studies, the optimal conditions established for the study of the Enterobacteriaceae biofilms required growth of the biofilm as a pellicle at the air-liquid interface. This limited the use of optic bottom plates and microfluidic devices in the studies. However, when biofilm formation conditions permit, the biofilms can be grown in microfluidic chambers or flow cells. The biofilms could then be washed, and beads introduced through the microfluidic device with minimal disruption of the biofilm.
Addition of beads during biofilm growth. We chose to add excess beads to the biofilms, and then remove unassociated beads by gentle washing to optimize the number of beads present during the analysis. In the viscous E. faecalis biofilms, it is possible that the beads disassociated and reassociated with during the 20-minute imaging time. If a low number of beads are added at different times during biofilm growth, it might be possible to trap the beads in the biofilm, allowing more accurate characterization of the biofilm movement in more viscous biofilms.
Choice of the region to image. For studies on material properties, it is best to choose thick and thin regions of the biofilm. However, when studying changes in the material properties of a treated biofilm, thick confluent regions may be imaged to determine changes in viscoelastic properties and bead penetration in those regions. In this case, looking for beads that penetrated the biofilm and ended up trapped against the coverslip are a useful measure of biofilm disruption.
Imaging under flow. Using optic glass flow cells or microfluidic devices, movement of beads or bacteria in a biofilm under flow can be imaged. This can be done in different ways. It can be done by injection of beads into the whole chamber followed by brief incubation to allow association of the beads with the biofilm. The unassociated beads can be removed by washing and the biofilm imaged with or without flow. Conversely, a small number of beads can be introduced into one side of the chamber and their movement through and in the biofilm can be tracked under flow. When using flow, caution will need to be used in choosing the Mosaic bead tracking settings (Step 4.5). In the current studies the dynamics setting was Brownian. MSD calculations confirmed that the movement was likely to be diffusive, making Brownian the appropriate setting.
Matrix staining. In the current studies, staining with Syto9 examines cellular density and not biofilm structure density. For example, the presence of amyloids likely increases the density of the matrix material of the biofilm. The dependence of movement on the amyloid density could be determined by using fluorescent matrix stains in lieu of Syto9.
Fluorescently labelled bacteria. Fluorescently labelled bacteria can be used to track movement of exogenous bacteria through biofilms (e.g., plasmid-containing bacteria). The challenge with fluorescently labelled bacteria, such as Enterococci, is that they form singles, diplococci, and short chains, which complicates the ability to accurately track the bacteria. This process would be easier if the bacteria have a single-cell morphology.
Limitations
Limitations in trajectory visualization and stitching.
One limitation of the method is trajectory visualization and stitching. Reconstructed and analyzed trajectories consist of x, y, z point coordinates, where subsequent points define the linear path between these points. Visualization of such piecewise linear trajectories can be achieved by various tools. One approach was to use Python and Jupyter notebooks together with the Python plugins, Pandas and Matplotlib. While it was possible to visualize individual existing trajectories in the Journal of Bacteriology article where this technique was originally published34, there were still significant limitations that are being addressed in future research.
Currently, the number of reconstructed trajectories is larger than the number of beads in the biofilm, meaning multiple trajectories may correspond to one bead. This can be caused by a weak confocal signal in one frame where Mosaic will terminate a trajectory and initiate a second. This may register as multiple shorter trajectories for one bead, especially in less viscous biofilms. Another cause for the large number of trajectories is the lack of trajectory stitching. Especially in E. faecalis optic bottom well biofilms, beads visually remain associated with the biofilm during imaging (Supplemental Video 2). However, there were no trajectories longer than 10 and over 90% of the trajectories had a lifespan of 5 frames or less (Figure 3D). If the software is used to analyze only trajectories above a defined length (e.g., when tracing cells that are capable of transferring plasmids), shorter trajectories can automatically be removed from the data set. However, there are other purposes for which stitching the trajectories may be very important. Finally, the inability to track rapid bead movement as a single trajectory could result in more trajectories in Enterobacteriaceae biofilms due to the rapid movement in the Z direction resulting in elliptical-shaped beads (Figure 2G). The possibility of stitching trajectories disrupted by rapid anisotropic movement will be important to study the effect of the curli amyloid matrix in Enterobacteriaceae.
Significance
A computational workflow was developed to study bead trajectories to compare the material properties of 3D biofilms. The workflow enables researchers to identify critical parameters that can be used in computational modeling of fluid dynamics in heterogeneous biofilms. With the help of this open-source bead evaluator, the effect bacterial amyloid curli on the material properties could be studied, showing increased biofilm matrix rigidity due to curli. In a more general context, the evaluator can be used to study changes in biofilm structure induced by biofilm treatment or different environmental conditions, such as flow. For example, the tool is being used to analyze the effect of monoclonal antibody treatment on the disruption of biofilm structures in collaboration with the Tükel laboratory (LKSOM Temple University). The bead evaluator toolbox is fully adaptable and extendable in a modular fashion using VRL-Studio to further enhance and extend its functions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Work in the GQ and BAB labs received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. The authors acknowledge Isaac Klapper, Ph.D (Department of Mathematics, Temple University) for helpful discussion and Çagla Tükel (Department of Microbiology and Immunology, Temple University) for Enterobacteriaceae expertise in the initial publication of containing this technique.
Materials
| Name | Company | Catalog Number | Comments |
| 96-well plates, No. 1.5 Uncoated Coverslip, 5 mm Glass Diameter | MatTek | P96G1.55F | |
| Fisherbrand Cover Glasses: Circles | Fisher Scientific | 12-293-232P | 1.5 optic glass coverslip |
| Invitrogen Syto 9 Green Fluorescent Nucleic Acid Stain | Invitrogen | S34854 | |
| Molecular Probes FluoSpheres Carboxylate-modified Microspheres, 1 um, crimson fluorescent (625/645) | Molecular Probes | F8816 |
References
- Huang, R., Li, M., Gregory, R. L. Bacterial interactions in dental biofilm. Virulence. 2, 435-444 (2011).
- Nadell, C. D., Drescher, K., Wingreen, N. S., Bassler, B. L. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME Journal. 9, 1700-1709 (2015).
- Billings, N., Birjiniuk, A., Samad, T. S., Doyle, P. S., Ribbeck, K. Material properties of biofilms - key methods for understanding permeability and mechanics. Reports on Progress in Physics. 78, 036601 (2015).
- Araújo, G. R. d. e. S., Viana, N. B., Gómez, F., Pontes, B., Frases, S. The mechanical properties of microbial surfaces and biofilms. The Cell Surface. 5, 100028 (2019).
- Tallawi, M., Opitz, M., Lieleg, O. Modulation of the mechanical properties of bacterial biofilms in response to environmental challenges. Biomaterials Science. 5, 887-900 (2017).
- Tursi, S. A., Tükel, &. #. 1. 9. 9. ;. Curli-Containing Enteric Biofilms Inside and Out, Matrix Composition, Immune Recognition, and Disease Implications. Microbiology and Molecular Biology Reviews. 82, (2018).
- Dueholm, M. S., Albertsen, M., Otzen, D., Nielsen, P. H. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One. 7 (12), 51274 (2012).
- Hung, C., et al. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio. 4, 00645 (2013).
- Kikuchi, T., Mizunoe, Y., Takade, A., Naito, S., Yoshida, S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiology and Immunology. 49, 875-884 (2005).
- Gallo, P. M., et al. Amyloid-DNA Composites of Bacterial Biofilms Stimulate Autoimmunity. Immunity. 42, 1171-1184 (2015).
- Serra, D. O., Richter, A. M., Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. Journal of Bacteriology. 195, 5540-5554 (2013).
- Oh, Y. J. Characterization of curli A production on living bacterial surfaces by scanning probe microscopy. Biophysical Journal. 103, 1666-1671 (2012).
- Lembré, P., Di Martino, P., Vendrely, C. Amyloid peptides derived from CsgA and FapC modify the viscoelastic properties of biofilm model matrices. Biofouling. 30, 415-426 (2014).
- Oh, Y. J., et al. Curli mediate bacterial adhesion to fibronectin via tensile multiple bonds. Scientific Reports. 6, 33909 (2016).
- Vidakovic, L., Singh, P. K., Hartmann, R., Nadell, C. D., Drescher, K. Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nature Microbiology. 3, 26-31 (2018).
- Tursi, S. A., et al. Salmonella Typhimurium biofilm disruption by a human antibody that binds a pan-amyloid epitope on curli. Nature Communications. 11, 1007 (2020).
- Perov, S., et al. Structural Insights into Curli CsgA Cross-β Fibril Architecture Inspire Repurposing of Anti-amyloid Compounds as Anti-biofilm Agents. PLOS Pathogens. 15 (8), 1007978 (2019).
- Taglialegna, A., et al. The biofilm-associated surface protein Esp of Enterococcus faecalis forms amyloid-like fibers. Npj Biofilms and Microbiomes. 6, 15 (2020).
- Gour, S., Kumar, V., Rana, M., Yadav, J. K. Pheromone peptide cOB1 from native Enterococcus faecalis forms amyloid-like structures, A new paradigm for peptide pheromones. Journal of Peptide Science. 25, 3178 (2019).
- Stoodley, P., Debeer, D., Lewandowski, Z. Liquid flow in biofilm systems. Applied and Environmental Microbiology. 60, 2711-2716 (1994).
- Birjiniuk, A., et al. Single particle tracking reveals spatial and dynamic organization of the E. coli biofilm matrix. New Journal of Physics. 16, 085014 (2014).
- Chew, S. C., et al. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio. 5, 01536 (2014).
- Cao, H., et al. Revealing region-specific biofilm viscoelastic properties by means of a micro-rheological approach. Npj Biofilms and Microbiomes. 2, 5 (2016).
- Galy, O., et al. Mapping of bacterial biofilm local mechanics by magnetic microparticle actuation. Biophysical Journal. 103, 1400-1408 (2012).
- Rogers, S. S., vander Walle, C., Waigh, T. A. Microrheology of bacterial biofilms in vitro, Staphylococcus aureus and Pseudomonas aeruginosa. Langmuir. 24, 13549-13555 (2008).
- Hart, J. W., Waigh, T. A., Lu, J. R., Roberts, I. S. Microrheology and Spatial Heterogeneity of Staphylococcus aureus Biofilms Modulated by Hydrodynamic Shear and Biofilm-Degrading Enzymes. Langmuir. 35 (9), 3553-3561 (2019).
- van Merode, A. E. J., van der Mei, H. C., Busscher, H. J., Krom, B. P. Influence of culture heterogeneity in cell surface charge on adhesion and biofilm formation by Enterococcus faecalis. Journal of Bacteriology. 188, 2421-2426 (2006).
- Tariq, M., Bruijs, C., Kok, J., Krom, B. P. Link between Culture Zeta Potential Homogeneity and Ebp in Enterococcus faecalis. Applied and Environmental Microbiology. 78, 2282-2288 (2012).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ, 25 years of image analysis. Nature Methods. 9, 671-675 (2012).
- . Oracle.com Available from: https://docs.oracle.com/en/java (2021)
- Hoffer, M., Poliwoda, C., Wittum, G. Visual reflection library, a framework for declarative GUI programming on the Java platform. Computing and Visualization in Science. 16, 181-192 (2013).
- Perov, S., et al. Structural Insights into Curli CsgA Cross-β Fibril Architecture Inspire Repurposing of Anti-amyloid Compounds as Anti-biofilm Agents. PLOS Pathogens. 15, 1007978 (2019).
- Malhotra, K., et al. Development of a New Bead Movement-Based Computational Framework Shows that Bacterial Amyloid Curli Reduces Bead Mobility in Biofilms. Journal of Bacteriology. 202, 00253 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved