Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Profiling of Pre-micro RNAs and microRNAs using Quantitative Real-time PCR (qPCR) Arrays
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
We will demonstrate the setup and analysis of pre-microRNA 96-well arrays for QPCR using a robot as well as by hand with a Thermo Scientific Matrix multichannel pipette.
Streszczenie
Protokół
The qPCR pre-miRNA profiling arrays can be set up as fully automated with a Tecan Freedom Evo robot (A) or by hand with the Matrix electronic multichannel pipette (B).
1)Prepare the master mix, primer plates and samples.
- Primer plates containing 186 primer pairs in 96-well format (total of 2 plates) at 0.5pM should be stored at -80°C. Thaw plates out at room temperature, vortex and centrifuge briefly before use.
- To prepare the master mix, thaw out SYBR Green 2x PCR Mix at room temperature. Each reaction uses 8ul master mix with 2ul primer. The composition of the master mix per reaction is 4ul SYBR mix, 3ul PCR grade water and 10-20 ng sample DNA or cDNA.
- For setup of each 96-well primer plate, four master mix tubes will be needed. Samples can be run 4 samples per plate (singlicate), 2 samples per plate (duplicate) or a single sample in quadruplicate.
- Prepare master mix by combining SYBR, water and sample in each of 4-2ml Eppendorf tubes. Each tube should contain enough master mix for approximately 100 reactions, allowing excess for pipetting waste. Vortex to mix.
A. Setup of pre-miRNA assay using the Freedom Tecan Evo robot.
- Following initialization of the robot and loading of the Evoware program, flush three times with 30mls each to clear system of air bubbles that will interfere with pipetting accuracy.
- Setup the robot platform to include the following:
- Labeled 384-well plate
- 96-well primer plate (1)
- Master mixes
- A trough containing 2% bleach
- A filled system fluid container
- Empty waste container
- Begin automated robot run by selecting "Run" twice.
- At end of program, remove 384-well plate and seal with LightCycler 480 sealing foil and centrifuge briefly. Place sealed 384-well plate in position 1 of the hotel.
- Go to step 2.2 and repeat for primer plate 2 with new master mixes and a new 384-well plate.
- Place sealed plate in position 2 of the hotel.
- Open new Evoware program for Loading the Lightcycler from the hotel and select "Run" twice.
- Freedom Evo will automatically load each plate into the LightCycler and run the following SYBR-green I/HRM cycling program:
Pre (1 cycle):
50° for 5 minutes at ramp rate of 4.8°/sec
95° for 5 minutes at ramp rate of 4.8°/sec
Amp (45 cycles):
95° for 15 sec at ramp rate of 4.8°/sec
62° for 30 sec at ramp rate of 2.5°/sec
(Single data acquisition during this step)
Melting curve:
95° for 5 sec at ramp rate of 4.8°/sec
60° for 1 minute at ramp rate of 2.5°/sec
95° continuous at ramp rate of .11°/sec
with 5 acquisitions per °C
Cool :
50° for 30 sec at ramp rate of 25°/sec
- Freedom Evo will automatically load each plate into the LightCycler and run the following SYBR-green I/HRM cycling program:
B.Setup of pre-miRNA assay using the Matrix electronic multichannel pipette:
- Place contents of master mix tube 1 into a reservoir.
- Set the electronic pipette to aspirate 16ul of master mix with 2-8ul dispense steps followed by a purge step.
- For master mix 1, dispense 8ul into every other well of the 384-well plate (set on 384-well tip spacing), beginning with well A1. (i.e. wells A1, C1, E1, etc.) For the second 8ul dispense, move to column 3 (i.e. wells A3, C3, E3, etc.) then purge over a waste container and dispose tips.
- Follow this pattern for the remaining 5 cycles until you reach well A23.
- Repeat for master mix 2, (beginning with well A2, C2, E2, etc.); master mix 3 (beginning at well B1, D1, F1, etc.); master mix 4 (beginning at B2, D2, F2, etc.).
- For aliquotting of primer plate, set electronic pipette to aspirate 2ul and dispense 2ul with a purge step following. Dispense 2ul of primers from column 1 of 96-well primer plate (A-H) into the 384-well plate, wells A1, C1, E1, etc. Purge over a waste container and dispose tips. Repeat for wells A2, C2, E2; B1, D1 ,F1; and B2, D2, F2, etc.
- Repeat for rows 2-12 of 96-well primers, moving over every 2 columns for each primer column, until full 384-well plate has been aliquotted.
- Seal plates with a LightCycler sealing foil and centrifuge briefly.
- Place plates into a LightCycler 480 and cycle the plates according to the following settings using the SYBR Green I/HRM detection format:
Pre (1 cycle):
50° for 5 minutes at ramp rate of 4.8°/sec
95° for 5 minutes at ramp rate of 4.8°/sec
Amp (45 cycles):
95° for 15 sec at ramp rate of 4.8°/sec
62° for 30 sec at ramp rate of 2.5°/sec
(Single data acquisition during this step)
Melting curve:
95° for 5 sec at ramp rate of 4.8°/sec
60° for 1 minute at ramp rate of 2.5°/sec
95° continuous at ramp rate of .11°/sec
with 5 acquisitions per °C
Cool :
50° for 30 sec at ramp rate of 25°/sec - Repeat using primer plate 2, aliquotting into a fresh 384-well plate using freshly prepared master mixes.
Secrets to success:
An important characteristic when designing PCR arrays is that all 186 primer pairs have the same annealing temperature, so the plate can be run at the same optimal PCR run program settings.
Each new array should be checked using three controls prior to running samples. These controls are:
1 run of all primers run with water or .1x TE to rule out primer contamination.
1 run of alternating water/TE and positive control, to ensure no carryover.
3 runs using the same positive control, to ensure reproducibility between runs.
Master mixes should be freshly prepared, and plates should be run within 24 hrs for optimal results
Primer plate foil should be removed slowly to reduce risk of contamination between wells.
When using a robot with fixed tips, 2% bleach wash is necessary to wash the tips between each pipetting step, followed by a water flush. This prevents and eliminates carryover, which could otherwise contaminate data and lead to inconclusive results.
After a plate is run through the Lightcycler, it should not be opened again in the PCR setup room. This helps prevent PCR contamination.
Representative Results:
qPCR results are typically represented by CT values as determined from the Lightcycler analysis software. A successful run usually consists of a range of CTs for samples, typically between 20-35 for samples that are positive. The water samples in a good run are always at a CT of >40 and any samples or specific wells with a CT over 37 are considered negative or not detected. As a general rule, samples yielding a CT of <10 are also unreliable and are excluded from further analysis. There are several ways to analyze qPCR data and the inclusion of internal controls in our assay helps control for variation of sample input. The pre-miR array includes a U6 control primer, which is often used as a reference gene to normalize CT values from different samples. This normalized value is referred to as the delta CT value (dCT). Results are often graphed as the raw CT, the dCT value or can be further analyzed to standardized values.
In Figure 1, analysis of the full pre-microRNA array of four different samples from a timecourse experiment is presented in heat map format. The relative expression of each pre-microRNA is shown and three major, distinct clusters emerged. The majority of pre-miRs analyzed underwent small, insignificant changes as expected (Figure 1A). However, a small portion of pre-microRNAs were significantly decreased (Figure 1B) or increased dramatically (Figure 1C) throughout the timecourse experiment. Levels of expression of each pre-microRNA were normalized to the 0h timepoint (sample 1) as a baseline level. In some instances, you may note that some of the clustered pre-microRNAs also cluster together in the heatmap analysis (ex: let-7a, Figure 1B). Depending on the experiment, clusters may emerge that are dependent on viral infection, cell type specific expression of pre-microRNAs or pre-miRs that are regulated by a common molecule or signaling pathway.
The pre-microRNA assay that we have developed also includes a number of known viral pre-microRNAs and genes encoded by Kaposi's Sarcoma Associated Herpesvirus (KSHV) and Epstein Barr Virus (EBV) in addition to the human pre-miRs. These can be used as positive or negative controls, depending on the cell line and viral status of the samples used. In Figure 2, the average CTs from a KSHV-negative sample run in quadruplicate are shown. Importantly, KSHV LANA was not detected in this sample by the highly sensitive qPCR array. However, U6--our internal control and let-7a--a highly expressed human pre-microRNA, were expressed at detectable levels. Finally, as expected, the negative control of PCR-grade water did not yield a qPCR product.
Another indicator of a successful qPCR run is a good melting curve analysis, which can give insight into potential contamination in samples. To establish a melting temperature, the plate is heated slowly after the PCR is complete. As the DNA strands separate, SYBR green is released, and fluorescence signal decreases. The temperature at which this drop occurs is graphed, and is known as the melting temperature of the sample. Depending on the DNA sequence, samples melt at different temperatures from each other and from the water negative control, which should not have a melting temperature since no DNA is present. The PCR products from each primer pair should melt at similar temperatures. For example, Figure 3 shows a melting curve analysis for two different primers using the same sample in duplicate. It is clear that the melting temperature is different for the two primer pairs but the sample is melting at the exact same temperature for each replicate. Signs of a bad or potentially contaminated sample may include several melting peaks, suggesting the presence of two different sources of input.
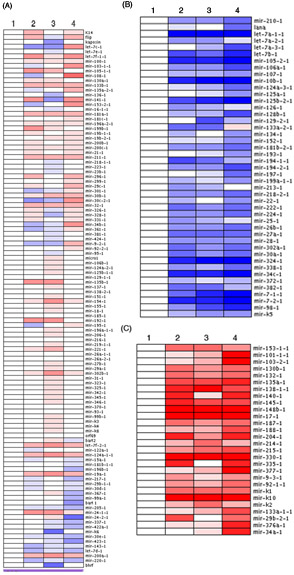
Figure 1. Pre-microRNA signatures emerge through profiling using a novel qPCR-based array. The average deltaCTU6 was calculated and standardized values were loaded into ArrayMinerTM software, yielding 3 distinct clusters displayed as heatmaps. (A) Pre-microRNAs with small changes in expression are shown. Pre-microRNAs whose levels were significantly decreased (B) or increased (C) are also shown. Blue corresponds to lower levels of expression while red represents increased expression levels.
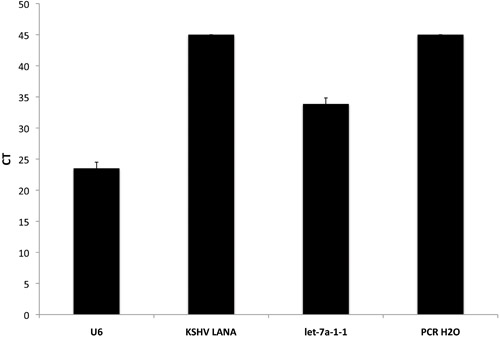
Figure 2. Inclusion of internal controls in the qPCR-based pre-microRNA array. The raw CT values for 3 different genes and a no template control are shown. U6 is used as an internal positive control while PCR H2O serves as our negative control. Let-7a-1-1 is a microRNA normally expressed in many different cell types while KSHV LANA can be used as either a positive or negative control, based on viral status of the cell line or tissue used.
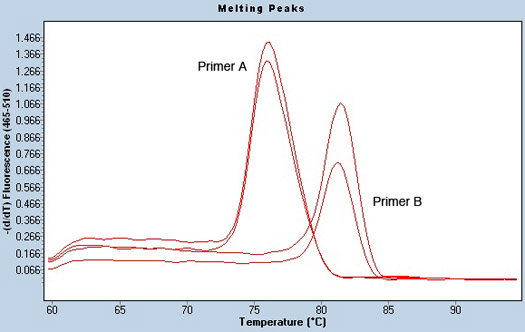
Figure 3. Melting peak analysis of pre-microRNA primers and lack of sample contamination. Melting curves were obtained using the Tm calling analysis in the LightCycler software. Two different primers (Primer A and B) for the same sample run in duplicate are shown. The melting temperature for each primer is distinct. However, the sample has only one peak, demonstrating the lack of sample contamination.
Access restricted. Please log in or start a trial to view this content.
Dyskusje
QPCR is a highly sensitive assay that can be used to compare gene expression levels between samples in a study, as well as to determine positivity in the case of viruses. The benefit of using qPCR arrays is the ability to run many primers (in our case, 186 primer pairs) for each sample in a short period of time. Using a pipetting robot such as the Tecan Freedom Evo, the amount of time required to set an experiment up can be further reduced, and the accuracy and consistency of the robot can reduce and eliminate pipettin...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
No conflicts of interest declared.
Podziękowania
This work was supported by NIH grants DE018304, R01DE018281. K.T. is supported by T32 GM07092-34 and by a grant to the University of North Carolina at Chapel Hill from Howard Hughes Medical Institute (HHMI) through the Med into Grad Initiative. PC is supported by T32 CA009156.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Freedom Evo 150 | Tecan Group Ltd. | 30017587 | |
| Matrix Electronic Multichannel Pipette | Thermo Fisher Scientific, Inc. | 2001-MTX | (or any comparable Thermo Scientific multichannel that can pipette the volumes described above) |
| Matrix Integrity Filter Tips, Sterile | Thermo Fisher Scientific, Inc. | 7435 | (or comparable tip for multichannel used) |
| Lightcycler 480 SYBR Green 1 Master | Roche Group | 04 707 516 001 | |
| Lightcycler 480 Instrument II | Roche Group | 05015243001 | 384-well version |
| Lightcycler 480 Multiwell Plate 384, white | Roche Group | 04729749001 | |
| LightCycler 480 Sealing Foil | Roche Group | 04729757001 | |
| LightCycler 480 Multiple Plate Analysis Software | Roche Group | 05075122001 | |
| Eliminase Decontaminant | Decon Laboratories | 04-355-31 |
Odniesienia
- Bustin, A. A. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chem. 55 (4), 611-622 (2009).
- Frégeau, C. J. Automated Processing of Forensic Casework Samples using Robotic Workstations Equipped with Nondisposable Tips - Contamination Prevention. J. Forensic Sci. 53 (3), 53-533 (2008).
- O'hara, A. J. Pre-micro RNA Signatures Delineate Stages of Endothelial Cell Transformation in Kaposi Sarcoma. PLoS Pathog. 5 (4), e1000389-e1000389 (2009).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone