Method Article
Arabidopsis thaliana Polar Glycerolipid Profiling by Thin Layer Chromatography (TLC) Coupled with Gas-Liquid Chromatography (GLC)
W tym Artykule
Podsumowanie
Composition of polar lipid extracts and the fatty acid composition of individual glycerolipids are determined in a simple and robust lipid profiling experiment. For this purpose, glycerolipids are isolated by thin layer chromatography and subjected to transmethylation of their acyl groups. Fatty acyl methylesters are quantified by gas-liquid chromatography.
Streszczenie
Biological membranes separate cells from the environment. From a single cell to multicellular plants and animals, glycerolipids, such as phosphatidylcholine or phosphatidylethanolamine, form bilayer membranes which act as both boundaries and interfaces for chemical exchange between cells and their surroundings. Unlike animals, plant cells have a special organelle for photosynthesis, the chloroplast. The intricate membrane system of the chloroplast contains unique glycerolipids, namely glycolipids lacking phosphorus: monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), sulfoquinovosyldiacylglycerol (SQDG)4. The roles of these lipids are beyond simply structural. These glycolipids and other glycerolipids were found in the crystal structures of photosystem I and II indicating the involvement of glycerolipids in photosynthesis8,11. During phosphate starvation, DGDG is transferred to extraplastidic membranes to compensate the loss of phospholipids9,12.
Much of our knowledge of the biosynthesis and function of these lipids has been derived from a combination of genetic and biochemical studies with Arabidopsis thaliana14. During these studies, a simple procedure for the analysis of polar lipids has been essential for the screening and analysis of lipid mutants and will be outlined in detail. A leaf lipid extract is first separated by thin layer chromatography (TLC) and glycerolipids are stained reversibly with iodine vapor. The individual lipids are scraped from the TLC plate and converted to fatty acyl methylesters (FAMEs), which are analyzed by gas-liquid chromatography coupled with flame ionization detection (FID-GLC) (Figure 1). This method has been proven to be a reliable tool for mutant screening. For example, the tgd1,2,3,4 endoplasmic reticulum-to-plastid lipid trafficking mutants were discovered based on the accumulation of an abnormal galactoglycerolipid: trigalactosyldiacylglycerol (TGDG) and a decrease in the relative amount of 18:3 (carbons : double bonds) fatty acyl groups in membrane lipids 3,13,18,20. This method is also applicable for determining enzymatic activities of proteins using lipids as substrate6.
Protokół
1. Lipid Extraction
- Begin lipid extraction by harvesting 30 mg 4-week-old Arabidopsis leaves from plants grown on agar solidified medium or soil and transfer them into 1.5 mL polypropylene reaction tubes. Fresh leaves can be flash frozen in liquid nitrogen and stored at -80 °C.
- Add 300 μL extraction solvent composed of methanol, chloroform and formic acid (20:10:1, v/v/v) to each sample. Shake vigorously (using a paint shaker or similar) for 5 minutes.
- Add 150 μL of 0.2 M phosphoric acid (H3PO4), 1 M potassium chloride (KCl) and vortex briefly.
- Centrifuge at 13,000xg at room temperature for 1 minute. Lipids dissolved in the lower chloroform phase will be spotted onto TLC plates.
2. Thin Layer Chromatography (TLC)15
- To prepare TLC plates, submerge a 20cmx20cm silica gel coated TLC plate with loading strip for 30 sec into 0.15 M ammonium sulfate ((NH4)2SO4) solution, After submerging for 30 seconds, dry the plate for at least 2 days in a covered container. During activation the sublimation of ammonium leaves behind sulfuric acid, which protonates phosphatidylglycerol necessary for its separation from other glycerolipids.
- On the day of experiment, activate TLC plates by baking in an oven at 120 °C for 2.5 hours.
- After cooling down the activated plates to room temperature, use a pencil to draw a straight line (1.5 cm from the edge of the plate) across the plate at the origin of the chromatogram.
- In a fume hood, slowly deliver 3 X 20 μL of lipid extract in the lower chloroform phase using a 20 μL pipette with 200 μL yellow plastic tips under a slow stream of N2. For this purpose, a Tygon Tubing is connected to the regulator of the N2 tank. Keep the spot smaller than 1 cm in diameter. Each plate can hold up to 10 samples (when subsequent GLC analysis is planned).
- As the lipid spots completely dry in the fume hood, prepare the developing solvent composed of acetone, toluene, water (91 mL: 30 mL: 7.5 mL). If the ambient relative air humidity is high, separation could be affected. In this case water should be reduced to give (91 mL: 30 mL: 7.0 mL) to achieve the desired separation.
- Pour 80 mL developing solvent into a sealable TLC developing chamber (L: H: W=27.0: 26.5: 7.0, cm/cm/cm) and place the plate into the tank with the sample end facing down. Seal the tank using the clamp. The solvent will ascend the plate and lipids will be separated. The development time is approximately 50 minutes at room temperature.
- When the solvent front has reached 1 cm from the top of the plate, carefully remove the plate from the tank and dry completely in the fume hood for approximately 10 minutes.
- Lipids separated by TLC can be either reversibly stained briefly with iodine for quantitative analysis or irreversibly stained with sulfuric acid or α-naphthol.
- Sulfuric acid charring: spray the plate with 50% sulfuric acid in water in a glass spray bottle in the fume hood and bake at 120 °C for 15 minutes (Figure 2A).
- α-naphthol staining for glycolipids: spray the plate with 2.4% (w/v) α-naphthol in 10% (v/v) sulfuric acid, 80% (v/v) ethanol and bake at 120 °C for 3-5 minutes until glycolipid bands are stained pink or purple (Figure 2B). Overtreatment will lead to charring of all lipids due to presence of sulfuric acid in the reagent.
- Iodine staining: in a fume hood, place the plate into a closed TLC tank with iodine crystals (in a tray on the bottom leading to saturation of the atmosphere with iodine vapor until lipids are visible). Don't expose the plate to iodine too long as iodine may covalently modify polyunsaturated fatty acids (Figure 2C). Alternatively, to avoid any oxidation of lipids, only standard lanes interspersed with sample lanes should be stained using a glass wool plugged Pasteur pipette with iodine crystals through which N2 is blown over individual standard lanes.
3. Fatty Acyl Methylester (FAME) Reaction16
- Remove silica surrounding identified lipid spots from the TLC plate with a razor blade. Scrape the lipid containing silica and transfer the silica powder using a funnel into a glass tube with a Teflon (PTFE)-lined screw cap.
- Add 1 mL 1 N hydrochloric acid (HCl) in anhydrous methanol to each sample by glass pipette.
- Add 100 μL 50 μg mL-1 pentadecanoic acid (15:0) using 200 μL pipette to each sample as internal standard using a 200 μL pipette with 200 μL yellow plastic tip. Keep a tube with only pentadecenoic acid in methanolic HCl as a control. Close glass tubes tightly with Teflon-lined caps.
- Incubate glass tubes in an 80 °C water bath for 25 minutes. Tubes need to be sealed so that the solvent does not evaporate.
- After tubes have cooled down, add 1 mL 0.9% sodium chloride followed by 1 mL hexane and vortex vigorously. Centrifuge samples at 1000xg for 3 minutes.
- In the fume hood, remove the hexane/upper layer of the sample with Pasteur pipette and place it into a new 13x100 mm glass tube.
- Evaporate hexane under a slow stream of N2 without drying completely.
- Dissolve the resulting fatty acyl methylesters s in 60 μL hexane. Transfer samples into autosampler vials and cap tightly. Samples can be stored at 4 °C for short term and -20 °C for a few days.
4. Gas-Liquid Chromatography (GLC)10
- Before beginning GLC, Ensure that the helium, hydrogen and air cylinders are filled.
- Sufficient hexane must be added to the solvent reservoir and the waste container must be empty. For fatty acyl methylesters separation, attach a DB-23 column to the machine.
- Place vials into the autosampler. Start the Chemstation software for GLC on the system computer.
- Set the inlet temperature at 250 °C with helium flow rate at 48.6 mL min-1 and the pressure at 21.93 psi. The split ratio is 30.0: 1.
- The oven temperature is set initially at 140 °C for 2 min and raised to 160 °C at a rate of 25 °C min-1. Then set the temperature to increase from 160 °C to 250 °C at a rate of 8 °C min-1 and hold at 250 °C for 4 min followed by a decrease to 140 °C at a rate of 38 °C min-1. One run takes approximately 21 minutes.
- The temperature of the flame ionization detector is 270 °C with a hydrogen flow rate of 30.0 mL min-1, air flow rate at 400 mL min-1 and helium flow rate at 30.0 mL min-1.
- Enter the number of vials and sample names in the run sequence table. Set the 10 μL injector to inject 2 μL sample per vial.
- When the instrument is ready, initiate the run sequence.
5. Representative Results:
Examples of irreversible staining of TLC-separated lipids from 4-week-old Arabidopsis seedlings are shown in Figure 2. The sulfuric acid stained lipids (Figure 2A) are charred and appear as brown spots. α-naphthol is preferred to stain glycolipids such as MGDG, DGDG, SQDG etc. Glycolipids stained with α-naphthol carry a pink-purple color while other polar lipids stain yellow (Figure 2B). The iodine staining is reversible and gives lipids a yellowish color that will disappear over a short time as iodine evaporates (Figure 2C). Briefly iodine stained lipids can be subjected to GLC analysis although unstained lipids are preferable to reduce break down of lipids.
If done successfully, distinctive signals representing different Fatty acyl methylester will be observed after GLC (Figure 3). Fatty acyl methylester with shorter carbon chain and fewer double bonds have shorter retention time using the DB-23 column. Fatty acyl methylester profiling is a sensitive tool to identify mutants with altered lipid composition. In Figure 4, the MGDG18:3 fatty acid molar ratio is decreased in the tgd4-1 mutant compared to the wild type18. By dividing the moles of Fatty acyl methylester for one lipid class with the moles of all lipid classes, the molar ratio of each lipid are calculated. For example, to calculate the molar ratio of MGDG:
(MGDG) mol% = ∑ [FAMEs(MGDG)] / ∑ [FAMEs(total)] x100%
The resulting molar ratios of each lipid class from both the wild type and the mutant can be compared. For instance, the tgd4-1 mutant has increased relative amounts of MGDG and PG but decreased amounts of DGDG and PE (Figure 5)18.
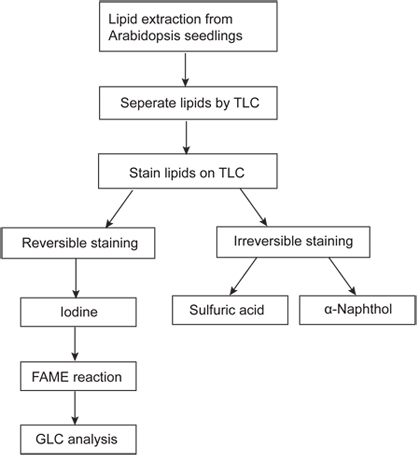
Figure 1. Flow chart of polar lipid analysis using Arabidopsis seedlings. Total lipids are extracted from 4-week-old Arabidopsis seedlings and separated by TLC. The separated lipids can be scraped from TLC plate for transesterification followed by GLC analysis.
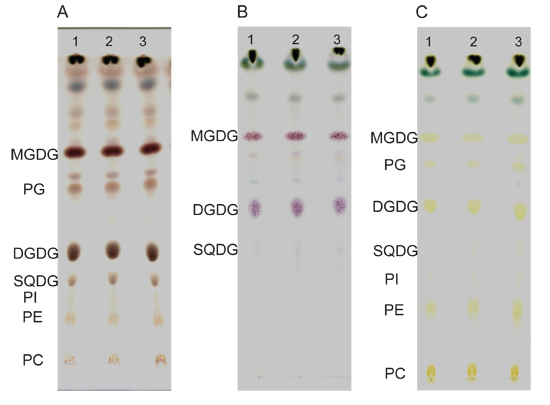
Figure 2. Separation of lipids on TLC plates. Lipid extracts of 35 mg (fresh weight) wild type seedlings are separated by TLC and stained by sulfuric acid (A), α-naphthol (B) or iodine vapor (C). Three repeats are shown in each staining method. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; SQDG, sulfoquinovosyldiacylglycerol.
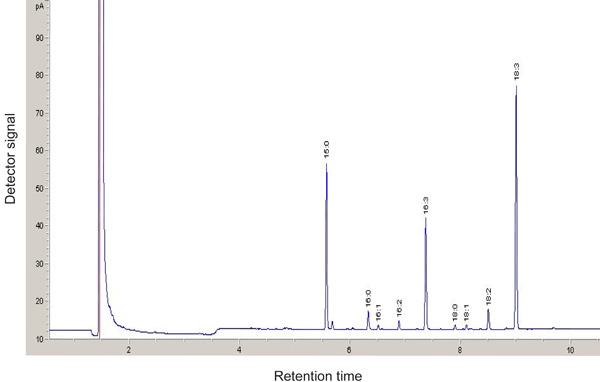
Figure 3. GLC analysis of Fatty Acid Methylesters (FAMEs) derived from MGDG of the wild type. FAMEs are separated on a 30 m capillary column and detected by flame ionization. Pentadecanoic acid (15:0) is used as an internal standard.

Figure 4. Fatty acid profile of MGDG in the wild type Col2 (white columns) and the tgd4-1 mutant (black columns). Fatty acids are presented as the number of carbons followed by the number of double bonds. Three repeats are averaged and standard deviations are shown.
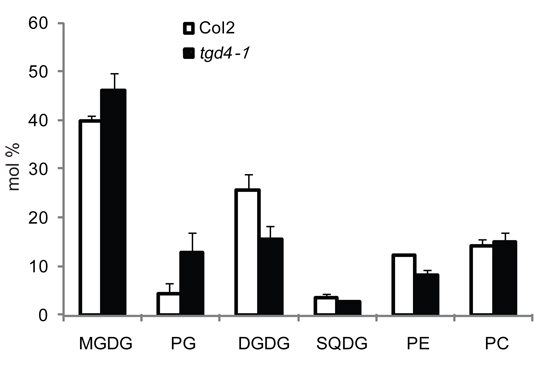
Figure 5. Polar lipids composition of the wild type Col2 (white columns) and the tgd4-1 mutant (black columns). Three repeats are averaged and standard deviations are shown by error bar.
Dyskusje
TLC coupled with GLC provides a robust and rapid tool for quantitative analysis of polar lipids in plants. Small changes in lipid composition can be identified; therefore, this method has been used for large scale screening of mutants impaired in polar lipid metabolic pathways1,20. This method is also widely used for monitoring activities of enzymes utilizing polar lipids as substrate.2,6,7
Besides leaves, the lipid composition of other plant tissues such as roots and seeds or subcellular fractions such as chloroplasts and mitochondria can also be determined in the same way.
The solvent system (acetone, toluene, water) used here is optimized for the separation of glycolipids and phospholipids in plants. However, in tgd1,2,3,4 mutants and isolated chloroplasts, TGDG runs together with PE while tetragalactosyldiacylglycerol runs with PC. In this case a solvent system with chloroform, methanol, acetic acid and water (85: 20: 10: 4, v/v/v/v) is used13. Sometimes two-dimensional TLC using two different solvent systems is performed to further separate glycolipids and phospholipids19. In addition, plant tissues can be directly subjected to the FAME reaction followed by GLC to determine the total fatty acid profile without initial separation on TLC5. Beside the demonstrated TLC-GLC system, another method used for lipid profiling is based on direct electrospray ionization tandem mass spectrometry17. In this method the initial chromatographic separation of lipids in the extract is omitted. However, this method requires expensive equipment and experienced personnel, which makes it less useful for routine analyses in the lab or for mutant screening.
Ujawnienia
No conflicts of interest declared.
Podziękowania
This work is supported by a grant from US National Science Foundation to Christoph Benning.
Materiały
| Name | Company | Catalog Number | Comments |
| α-naphthol | Sigma-Aldrich | N1000 | |
| Methanolic HCL 3N | Sigma-Aldrich | 33050-U | Dilute to 1N by methanol |
| Si250-PA TLC plates | JT Baker | 7003-04 | With pre-absorbent |
| TLC chamber | Sigma-Aldrich | Z266000 | |
| Screw cap tubes | VWR international | 53283-800 | |
| Scew caps | Sun Sri | 13-425 | |
| PTFE disk | Sun Sri | 200 608 | |
| GLC system | Hewlett-Packard | HP6890 | |
| DB-23 column | J&W Scientific | 122-2332 | |
| GLC vials | Sun Sri | 500 132 | |
| Caps of GLC vials | Sun Sri | 201 828 | |
| Chemstation software | Agilent Technologies | G2070AA |
Odniesienia
- Ajjawi, I. Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol. 152, 529-529 (2010).
- Andersson, M. X., Kjellberg, J. M., Sandelius, A. S. The involvement of cytosolic lipases in converting phosphatidyl choline to substrate for galactolipid synthesis in the chloroplast envelope. Biochim. Biophys. Acta. 1684, 46-46 (2004).
- Awai, K. A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. U. S. A. 103, e10817-e10817 (2006).
- Benning, C. Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71-71 (2009).
- Browse, J., McCourt, P. J., Somerville, C. R. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152, 141-141 (1986).
- Dormann, P., Balbo, I., Benning, C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science. 284, 2181-2181 (1999).
- Guskov, A. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. 16, 334-334 (2009).
- Hartel, H., Dormann, P., Benning, C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 97, 10649-10649 (2000).
- James, A. T., MARTIN, A. J. Gas-liquid partition chromatography; the separation and micro-estimation of volatile fatty acids from formic acid to dodecanoic acid. Biochem. J. 50, 679-679 (1952).
- Jordan, P. Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature. 411, 909-909 (2001).
- Kobayashi, K. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 57, 322-322 (2009).
- Lu, B. A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J. Biol. Chem. 282, 35945-35945 (2007).
- Ohlrogge, J., Browse, J. Lipid biosynthesis. Plant Cell. 7, 957-957 (1995).
- Stahl, E. Thin-layer chromatography; methods, influencing factors and an example of its use. Pharmazie. 11, 633-633 (1956).
- Stoffel, W., INSULL, W., AHRENS, E. H. Gas-liquid chromatography of highly unsaturated fatty acid methyl esters. Proc. Soc. Exp. Biol. Med. 99, 238-238 (1958).
- Welti, R., Wang, X., Williams, T. D. Electrospray ionization tandem mass spectrometry scan modes for plant chloroplast lipids. Anal. Biochem. 314, 149-149 (2003).
- Xu, C. Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell. 20, 2190-2190 (2008).
- Xu, C. Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell. 17, 3094-3094 (2005).
- Xu, C. A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 22, 2370-2370 (2003).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone