Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Study of the DNA Damage Checkpoint using Xenopus Egg Extracts
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
Xenopus egg extract is a useful model system to investigate the DNA damage checkpoint. This protocol is for the preparation of Xenopus egg extracts and DNA damage checkpoint inducing reagents. These techniques are adaptable to a variety of DNA damaging approaches in the study of the DNA damage checkpoint signaling.
Streszczenie
On a daily basis, cells are subjected to a variety of endogenous and environmental insults. To combat these insults, cells have evolved DNA damage checkpoint signaling as a surveillance mechanism to sense DNA damage and direct cellular responses to DNA damage. There are several groups of proteins called sensors, transducers and effectors involved in DNA damage checkpoint signaling (Figure 1). In this complex signaling pathway, ATR (ATM and Rad3-related) is one of the major kinases that can respond to DNA damage and replication stress. Activated ATR can phosphorylate its downstream substrates such as Chk1 (Checkpoint kinase 1). Consequently, phosphorylated and activated Chk1 leads to many downstream effects in the DNA damage checkpoint including cell cycle arrest, transcription activation, DNA damage repair, and apoptosis or senescence (Figure 1). When DNA is damaged, failing to activate the DNA damage checkpoint results in unrepaired damage and, subsequently, genomic instability. The study of the DNA damage checkpoint will elucidate how cells maintain genomic integrity and provide a better understanding of how human diseases, such as cancer, develop.
Xenopus laevis egg extracts are emerging as a powerful cell-free extract model system in DNA damage checkpoint research. Low-speed extract (LSE) was initially described by the Masui group1. The addition of demembranated sperm chromatin to LSE results in nuclei formation where DNA is replicated in a semiconservative fashion once per cell cycle.
The ATR/Chk1-mediated checkpoint signaling pathway is triggered by DNA damage or replication stress 2. Two methods are currently used to induce the DNA damage checkpoint: DNA damaging approaches and DNA damage-mimicking structures 3. DNA damage can be induced by ultraviolet (UV) irradiation, γ-irradiation, methyl methanesulfonate (MMS), mitomycin C (MMC), 4-nitroquinoline-1-oxide (4-NQO), or aphidicolin3, 4. MMS is an alkylating agent that inhibits DNA replication and activates the ATR/Chk1-mediated DNA damage checkpoint 4-7. UV irradiation also triggers the ATR/Chk1-dependent DNA damage checkpoint 8. The DNA damage-mimicking structure AT70 is an annealed complex of two oligonucleotides poly-(dA)70 and poly-(dT)70. The AT70 system was developed in Bill Dunphy's laboratory and is widely used to induce ATR/Chk1 checkpoint signaling 9-12.
Here, we describe protocols (1) to prepare cell-free egg extracts (LSE), (2) to treat Xenopus sperm chromatin with two different DNA damaging approaches (MMS and UV), (3) to prepare the DNA damage-mimicking structure AT70, and (4) to trigger the ATR/Chk1-mediated DNA damage checkpoint in LSE with damaged sperm chromatin or a DNA damage-mimicking structure.
Protokół
1. LSE Preparation
- Female frogs (Xenopus laevis) are injected twice for egg collection. The first injection (priming) is 100 U PMSG (Pregnant Mare Serum Gonadotropin) per frog. Frogs must be primed at least two days before inducing egg laying and primed frogs are usable for up to two weeks. To prime frogs, inject PMSG subcutaneously in the dorsal lymph sacs using a 3 ml syringe and 27 G needle.
- To induce egg laying, inject 500 U hCG (human Chorionic Gonadotrophin) per primed frog subcutaneously in the dorsal lymph sacs using a 27 G needle. Incubate injected frogs in separate buckets containing 2 liters of 1x Marc's modified Ringer's solution (MMR). Allow frogs 14-20 hr to lay eggs before collection.
- Remove the frogs from the buckets and pour off the MMR solution until around 100 ml is left. Obtain eggs from the buckets by transferring eggs to a 250 ml beaker.
- Dejelly eggs by adding 100 ml of 2% cysteine (adjust pH to 7.8 with 10 M KOH). Gently swirl the eggs with an inverted glass Pasteur pipet (0.7 cm in diameter) approximately every 30 sec. Decant and replace with fresh cysteine twice during incubation. The dejellying process is complete in about 5-15 min when the eggs form a more condensed layer at the bottom of the beaker.
- Discard the cysteine solution and wash eggs three times with 0.25x MMR. Swirl the eggs in the solution by a glass pipet. "Bad" eggs are determined by simple visual inspection and are "puffy" white in appearance. The "bad" eggs will accumulate at the center of the beaker after swirling. Remove "bad" eggs by a Pasteur pipet.
- Wash eggs with Egg Lysis Buffer (ELB) three times. Remove any additional "bad" eggs by a Pasteur pipet. Pour eggs into a 14 ml Falcon tube.
- Spin the Falcon tube for 55 sec at 188 x g (1,100 rpm) using the CL2 IEC clinical table-top centrifuge with a swinging bucket rotor to compact the eggs. Remove excess buffer from the top of the egg layer. Add 0.5 μl of Aprotinin/Leupeptin stock and 0.5 μl of Cytochalasin B stock per ml of compacted eggs.
- Centrifuge eggs at 16,500 x g (10,000 rpm) for 15 min at 4 °C using the Sorvall RC6 plus superspeed centrifuge with HB6 swinging bucket rotor. After centrifugation, eggs are fractionated into three layers in the tube: lipid, extract, and yolk/pigment from top to bottom, respectively. Puncture the side of the Falcon tube in the lower portion of the middle extract layer with a 21 G needle. Carefully remove the needle as the puncturing needle may be obstructed by plastic. Insert a fresh 21 G needle attached to a 1 ml syringe into the puncture site to collect the extract. Slowly aspirate the extract into syringe to avoid air bubbles and contamination with the lipid and yolk/pigment layers.
- Place the extracts into a chilled 1.5 ml microcentrifuge tube. For each milliliter of egg extract, add the following chemical stock solutions to the indicated final concentration (shown in parentheses): (1) 10 μl Cycloheximide (100 μg/ml); (2) 1 μl Aprotinin/Leupeptin (10 μg/ml each); (3) 1 μl Cytochalasin B (5 μg/ml); (4) 1 μl Dithiothreitol (1 mM); and (5) 0.33 μl Nocodazole (3 μg/ml). Invert the egg extract at least 10 times gently. This is the LSE, which must be made fresh and used within 4 hr. The quality of LSE is compromised after 4 hr or freeze-thaw.
2. Treatment of Sperm Chromatin with DNA Damaging Approaches
- Prepare normal sperm chromatin according to the method described previously 13.
- Prepare MMS-treated sperm chromatin
- Resuspend normal sperm chromatin in 0.5 ml Buffer X.
- Add ~ 5.5 μl of MMS (9.1 M in stock) to 500 μl resuspended chromatin to a final concentration of 100 mM.
- Incubate sperm chromatin in microcentrifuge tube at room temperature for 30 min with rotation.
- Spin the microcentrifuge tube at 686 x g (2,100 rpm) for 10 min at room temperature using the IEC CL2 clinical centrifuge with swinging bucket rotor and tube adaptors.
- Discard supernatant and resuspend the pellet in 0.5 ml of Buffer X plus BSA (3%), DTT (0.1 mM) and Aprotinin/Leupeptin (10 μg/ml each).
- Repeat steps (4) and (5) twice to wash the sperm chromatin.
- Determine the concentration of sperm chromatin with a hemocytometer and dilute to 100,000 sperm/μl. Save 5 μl aliquots in -80 °C freezer for further use.
- Prepare UV-treated sperm chromatin
- Add desired amount (e.g. 10 μl) of normal sperm chromatin on the surface of a piece of Parafilm.
- Place the Parafilm into a UV crosslinker. Set desired energy parameter, depending on your experiment, and start to damage the sperm chromatin via UV light. For example, it takes approximately 21 sec to reach 1,000 J/m2.
- After UV irradiation, UV-treated sperm chromatin should be added to egg extracts immediately.
3. Preparation of a DNA Damage-mimicking Structure (AT70)
- Dissolve two synthetic oligonucleotides poly-(dA)70 (designated as A70) and poly-(dT)70 (designated as T70) in water to a concentration of 2 μg/μl, respectively.
- Add 100 μl of A70 and 100 μl of T70 to one 1.5 ml microcentrifuge tube. Boil mixed synthetic oligo solution for 5 min at 95 °C in a heatblock.
- Take the dry bath block out (with the tube containing AT mixture in it), and let it cool down to room temperature on a lab bench. This temperature adjustment takes around 45-60 min.
- The cooled mixture is AT70 with a final concentration of 2 μg/μl. Store 10 μl of AT70 aliquots in -20 °C freezer for further use.
4. Triggering the DNA Damage Checkpoint in LSE with Damaged Sperm Chromatin or a DNA Damage-mimicking Structure
- Induce DNA damage checkpoint in LSE with damaged sperm chromatin
- Add 50 μl of LSE to a 1.5 ml microcentrifuge tube and complement it with 1 μl of Energy Mixture and 2 μl of damaged sperm chromatin (final concentration ~4,000 sperm/μl reaction).
- Incubate the reaction tube at room temperature for 90 min and flick the tube every 10 min.
- Dispense 1 μl of reaction mixture onto a microscope slide supplemented with 1 μl of nuclear dye solution after 30-min incubation. Place a cover slip over the reaction mixture and check for nuclei formation via fluorescence microscope. Typically, round nuclei will form after 30 min of incubation, indicating DNA replication has initiated.
- Take 10 μl of the reaction mixture into 90 μl of sample buffer. Samples are analyzed via immunoblotting using anti-Chk1 P-S344 or anti-Chk1 antibodies.
- Induce DNA damage checkpoint in LSE with the AT70
- Add 50 μl of LSE to a 1.5 ml microcentrifuge tube supplemented with 1 μl of Energy Mixture and 1.6 μl of Tautomycin stock.
- Add 1.25 μl of pre-made AT70 (2 μg/μl) or water (as negative control) to each reaction. Incubate the reactions at room temperature for 90 min. Flick the reaction tubes every 10 min.
- Add 10 μl of the reaction mixture into 90 μl of sample buffer. Samples are examined via immunoblotting using anti-Chk1 P-S344 or anti-Chk1 antibodies.
5. Representative Results
The damaged sperm chromatin or DNA damage-mimicking structure can trigger the ATR/Chk1-mediated DNA damage checkpoint in the Xenopus egg extract system. Figure 2A shows that MMS induces Chk1 phosphorylation at Ser344 (Chk1 P-S344), which is an indicator of ATR kinase activation. Figure 2B shows that AT70, as a DNA damage-mimicking structure, also triggers Chk1 phosphorylation. Total Chk1 samples are used as loading controls in both examples.
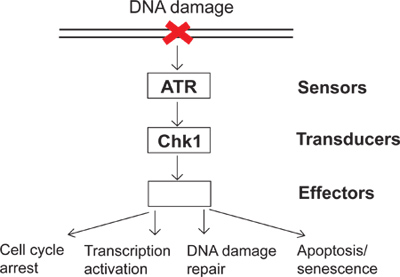
Figure 1. A diagram of the DNA damage checkpoint signaling.
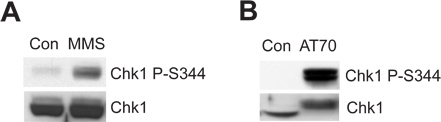
Figure 2. Chk1 phosphorylation is induced by either MMS or AT70 treatments in Xenopus egg extracts. (A) MMS-damaged sperm chromatin (MMS) or normal sperm chromatin (Con) are incubated in egg extracts for 90 min. Chk1 phosphorylation at Ser344 (Chk1 P-S344) and total Chk1 in egg extracts are examined via immunoblotting. (B) AT70 or water (Con) are added into egg extracts, respectively. Samples are also analyzed via immunoblotting as in (A).
Access restricted. Please log in or start a trial to view this content.
Dyskusje
There are several advantages in studying the DNA damage checkpoint using Xenopus egg extracts. The use of egg extracts provides a large quantity of cell-free extracts synchronized at interphase of the cell cycle. The egg extracts can be easily and inexpensively made. It is relatively easy to damage DNA or chromatin and to reveal a defect in the DNA damage checkpoint after immunodepleting a target protein from egg extract. Subsequently, a potential function defect can be "rescued" by addback of wild type or muta...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
No conflicts of interest declared.
Podziękowania
This work is supported in part by funds provided by The University of North Carolina at Charlotte, Wachovia foundation fund for faculty excellence, and a grant from NIGMS (R15GM101571).
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Reagents | |||
| Anti-Chk1 P-S344 antibody | Cell Signaling | 2348L | |
| Anti-Chk1 antibody | Santa Cruz | SC7898 | |
| Aprotinin | MP Biomedicals | 0219115880 | |
| Cycloheximide | Sigma | C7698-5G | |
| Cytochalasin B | EMD | 250233 | |
| Dithiothreitol (DTT) | VWR | JTF780-2 | |
| hCG | Sigma | CG10-10VL | |
| L-Cysteine | Sigma | C7352-1KG | |
| Leupeptin | VWR | 97063-922 | |
| Methyl methanesulfonate (MMS) | Sigma | 129925-5G | |
| Nocodazole | Sigma | M1404-2MG | |
| PMSG | Calbiochem | 367222 | |
| Sample buffer | Sigma | S3401 | |
| Tautomycin | Wako Chemicals USA | 209-12041 | |
| Equipment | |||
| Bucket for egg laying | Rubbermaid commercial products | 6308 | |
| CL2 IEC centrifuge with swinging bucket rotor | Thermo Scientific | 004260F | |
| HB6 swinging bucket rotor | Thermo Scientific | 11860 | |
| Sorvall RC6 plus superspeed centrifuge | Thermo Scientific | 46910 | |
| UV crosslinker | UVP | 95-0174-01 | |
| Solutions | |||
| 1x MMR | 100 mM NaCl, 2 mM KCl, 0.5 mM MgSO4, 2.5 mM CaCl2, 5 mM HEPES, adjust pH to 7.8 with 10 M NaOH | ||
| Aprotinin/Leupeptin stock | 10 mg/ml each in water. Store 20 μl aliquots at -80 °C. | ||
| Buffer X | 0.2 M sucrose, 80 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10 mM HEPES, adjust pH to 7.5 by HCl | ||
| Cycloheximide stock | 10 mg/ml in water. Store 1 ml aliquots at -20 °C. | ||
| Cytochalasin B stock | 5 mg/ml in DMSO. Store 20 μl aliquots at -20 °C. | ||
| Dithiothreitol (DTT) stock | 1 M in water. Store 1 ml aliquots at -20 °C. | ||
| ELB | 0.25 M sucrose, 1 mM DTT, 50 μg/ml cycloheximide, 2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES, pH7.7 | ||
| Nocodazole stock | 10 mg/ml in DMSO. Store 5 μl aliquots at -80 °C. | ||
| Energy Mixture | 375 mM creatine phosphate, 50 mM ATP, and 25 mM MgCl2. Aliquots are saved at -80 °C. | ||
| Nuclear dye solution | 0.4 μg/ml H–chst 33258, 25% glycerol (v/v), in 1x PBS | ||
| Tautomycin stock | 100 μM in DMSO. Store 10 μl aliquots at -80 °C. | ||
Odniesienia
- Lohka, M. J., Masui, Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 220 (4598), 719-721 (1983).
- Cimprich, K. A., Cortez, D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9 (8), 616-627 (2008).
- Lupardus, P. J., Van, C., Cimprich, K. A. Analyzing the ATR-mediated checkpoint using Xenopus egg extracts. Methods. 41 (2), 222-231 (2007).
- Lupardus, P. J., Byun, T., Yee, M. C., Hekmat-Nejad, M., Cimprich, K. A. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 16 (18), 2327-2332 (2002).
- Stokes, M. P., Van Hatten, R., Lindsay, H. D., Michael, W. M. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 158 (5), 863-872 (2002).
- Kato, K., Strauss, B. Accumulation of an intermediate in DNA synthesis by HEp.2 cells treated with methyl methanesulfonate. Proc. Natl. Acad. Sci. U.S.A. 71 (5), 1969-1973 (1974).
- Paulovich, A. G., Hartwell, L. H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 82 (5), 841-847 (1995).
- Guo, Z., Kumagai, A., Wang, S. X., Dunphy, W. G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14 (21), 2745-2756 (2000).
- Jazayeri, A., Balestrini, A., Garner, E., Haber, J. E., Costanzo, V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO. J. 27 (14), 1953-1962 (2008).
- Kumagai, A., Dunphy, W. G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 6 (4), 839-849 (2000).
- Yan, S., Lindsay, H. D., Michael, W. M. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 173 (2), 181-186 (2006).
- Shiotani, B., Zou, L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 33 (5), 547-558 (2009).
- Tutter, A. V., Walter, J. C. Chromosomal DNA replication in a soluble cell-free system derived from Xenopus eggs. Methods Mol. Biol. 322, 121-137 (2006).
- Cross, M. K., Powers, M. Preparation and Fractionation of Xenopus laevis Egg Extracts. J. Vis. Exp. (18), e891(2008).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone