Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
3D Whole-heart Myocardial Tissue Analysis
* Wspomniani autorzy wnieśli do projektu równy wkład.
W tym Artykule
Podsumowanie
This protocol describes a novel method for the 3D comparison of whole-heart myocardial tissue with MRI. This is designed for the accurate assessment of intramyocardial injections in the infarct border zone of a chronic porcine model of myocardial infarction.
Streszczenie
Cardiac regenerative therapies aim to protect and repair the injured heart in patients with ischemic heart disease. By injecting stem cells or other biologicals that enhance angio- or vasculogenesis into the infarct border zone (IBZ), tissue perfusion is improved, and the myocardium can be protected from further damage. For maximum therapeutic effect, it is hypothesized that the regenerative substance is best delivered to the IBZ. This requires accurate injections and has led to the development of new injection techniques. To validate these new techniques, we have designed a validation protocol based on myocardial tissue analysis. This protocol includes whole-heart myocardial tissue processing that enables detailed two-dimensional (2D) and three-dimensional (3D) analysis of the cardiac anatomy and intramyocardial injections. In a pig, myocardial infarction was created by a 90-min occlusion of the left anterior descending coronary artery. Four weeks later, a mixture of a hydrogel with superparamagnetic iron oxide particles (SPIOs) and fluorescent beads was injected in the IBZ using a minimally-invasive endocardial approach. 1 h after the injection procedure, the pig was euthanized, and the heart was excised and embedded in agarose (agar). After the solidification of the agar, magnetic resonance imaging (MRI), slicing of the heart, and fluorescence imaging were performed. After image post-processing, 3D analysis was performed to assess the IBZ targeting accuracy. This protocol provides a structured and reproducible method for the assessment of the targeting accuracy of intramyocardial injections into the IBZ. The protocol can be easily used when the processing of scar tissue and/or validation of the injection accuracy of the whole heart is desired.
Wprowadzenie
Ischemic heart disease has been the world's leading cause of death for the past decades1. Acute treatment after myocardial infarction aims to restore blood flow to the myocardium via percutaneous coronary intervention or coronary artery bypass grafting. In severe infarctions, a large area of the myocardium is scarred, and these cases often result in ischemic heart failure (HF)2. Current treatment options for HF focus on prevention and the preservation of cardiac function for the HF patients, but not on regeneration.
In the last decade, cardiac regenerative therapies have been investigated as a treatment option for HF3. This therapy aims to deliver biologicals, such as stem cells or growth factors, directly to the injured myocardium to induce revascularization, cardiomyocyte protection, differentiation, and growth4. For optimal therapeutic effect, it is hypothesized that the biological must be injected in the infarct border zone (IBZ) to facilitate good tissue perfusion for the survival of the biological and for optimal effect to the target zone5,6. Multiple techniques have been developed to perform identification and visualization of the IBZ to guide intramyocardial injections7,8,9,10,11. Besides identification and visualization of the IBZ, the delivery also relies on the biomaterials and injection catheters used. To validate the injection accuracy of the delivery techniques, an accurate and reproducible quantification method is required.
We have developed a protocol for whole-heart myocardial tissue processing that offers two-dimensional (2D) and three-dimensional (3D) imaging, which can be used for qualitative and quantitative study aims. The protocol covers the embedding process and the digital image analysis. In this paper, we demonstrate a protocol for the assessment of the targeting accuracy of intramyocardial injections in the IBZ in a large porcine model of chronic myocardial infarction.
Protokół
The in vivo experiment was conducted in accordance with the Guide for the Care and Use of Laboratory Animals prepared by the Institute of Laboratory Animal Research. The experiment was approved by the local Animal Experiments Committee.
1. Preparation of Injectable and Embedding Solution
- Prepare the injectable gel.
- Prepare 1 mL of ureido-pyrimidinone (UPy) gel in accordance to previously-described protocols12,13.
- Add superparamagnetic iron oxide particles (SPIOs) to the solution to get a concentration of 15 µg/mL and stir the mixture for 5 min for uniform distribution.
- Add the fluorescent microbeads to the solution to get a concentration of 10,000 beads/mL and stir the mixture for 5 min for uniform distribution.
- Store the resulting mixture at room temperature in a dark environment. Warm and vortex or stir the solution shortly before the injection procedure.
- Prepare the embedding solution.
- Start with tap water at room temperature and add agarose (agar) to a concentration of 4 wt%.
- Slowly heat up the solution to the boiling point using a microwave oven and stir frequently during heating. Upon reaching the boiling point, store and keep the agar solution above 70 °C for 2 h to allow trapped air to surface.
- Allow the agar to cool down at room temperature to a temperature between 50 and 60 °C until the time of embedding.
2. Injection Procedure
- Perform premedication (anti-arrhythmic agents, anti-platelet therapy, and pain medication), anesthesia, venous access, and intubation, as previously described14.
- Perform injections using an intramyocardial injection catheter (Table of Materials). For each injection, 0.2 mL of the mixture is injected in one bolus at a constant rate of approximately 0.3 mL/min using an injection device. Place the injections at different positions along the IBZ12.
- Administer 0.2 mL/kg (1.0 mmol/mL) of a gadolinium-based contrast agent 15 min prior to euthanizing the animal.
- Administer 20 mL of 7.5% potassium chloride intravenously to euthanize the animal.
- Secure mediastinal access following protocol steps 8.2 - 8.3, as described by Koudstaal et al.14. Cut the inferior caval vein 5 cm from the right atrium and remove outflowing blood with a suction device. Excise the heart and rinse it with 0.9% saline at room temperature.
3. Embedding Procedure
- Prepare the heart.
- Remove the pericardium from the heart while keeping the atria and ventricles intact. Dissect the ascending aorta ± 1 cm above the aortic valve using Klinkenberg scissors. Cut the inferior caval vein ± 1 cm from the atrium, and do the same for the pulmonary veins.
- Suture the apex of the heart to the bottom of a plastic embedding container (17 x 15 x 15 cm, W x D x H) using a 2-0 suture to prevent floatation of the heart during embedding (Figure 1A).
- Suture the remaining part of the aorta to the rims of the container using 2-0, making sure that the heart is centered and not touching the walls of the container (Figure 1B).
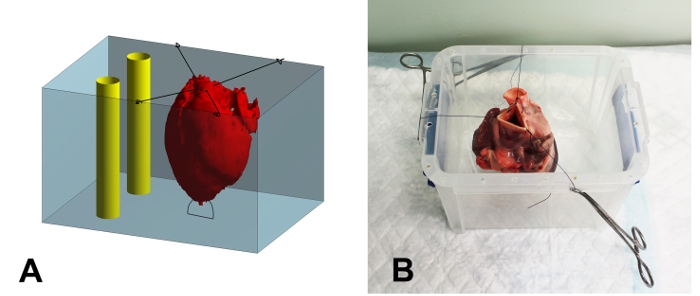
Figure 1: Schematic Overview and Photograph of the Embedding Container. (A) Schematic overview of the embedding process. The heart (red) is secured in the container (blue) using sutures. After filling the heart with the agar solution, the space around the heart is filled. Finally, two rigid plastic tubes (yellow) are positioned in the container, next to but not touching the heart, to serve as a reference during image registration. (B) Photograph of a heart secured in the embedding container. The sutures are clamped to the rim of the container using mosquito clamps. Please click here to view a larger version of this figure.
- Embed the heart in an end-diastolic-like geometry.
NOTE: The prevention of air bubble creation is necessary. If large air bubbles present in the agar solution, keep the agar at 40 °C, allowing the air bubbles to surface.- Clamp the inferior caval vein using mosquito clamps. Slowly inject the liquid agar using a 50-mL syringe in the right atrium via the superior caval vein until both the right atrium and ventricle are completely filled.
- Clamp the pulmonary veins using mosquito clamps. Gently pass an agar-filled 50-mL syringe retrogradely through the aortic valves. Slowly inject the solution in the left ventricle (LV) until the LV and the left atrium are completely filled. After filling the LV, clamp the aorta to keep the agar in the LV.
- Pour the remaining agar into the container until the heart is fully covered. Place two rigid plastic tubes within the embedding container to serve as reference structures for later image registration (Figure 1A). Make sure the tubes do not touch the walls of the container or the heart.
- Let the agar solidify at 2 - 7 °C.
4. Image Acquisition
- Perform transversal ex vivo MRI scans of the heart that is embedded in the container.
- Place the container with the embedded heart inside a head coil (Table of Materials).
- Angulate the slices parallel to the bottom of the container. Use the same orientation and angulation in each ex vivo MRI sequence.
- To visualize myocardium, perform a fluid-attenuated inverse recovery (FLAIR) sequence with the following parameters: repetition time [TR]/echo time [TE] = 10 s/140 ms, flip angle = 90°, pixel size = 0.5 x 0.5 mm, field of view [FOV] = 169 x 169 mm, 320 x 320 matrix, and 3-mm slice thickness.
- To visualize the myocardial infarction, perform a late-gadolinium enhanced (LGE) sequence with the following parameters: [TR]/[TE] = 5.53 ms/1.69 ms, flip angle = 25°, pixel size = 1.0 x 1.0 mm, [FOV] = 169 x 169 mm, 176 x 176 matrix, and 3-mm slice thickness.
- To visualize SPIOs, perform a T2*-weighted gradient echo sequence with the following parameters: [TR]/[TE] = 88.7 ms/15 equally-distributed TEs with a range of 1.9 - 24.6 ms, flip angle = 15°, pixel size = 0.5 x 0.5 mm, [FOV] = 169 x 169 mm, 320 x 320 matrix, and 3-mm slice thickness.
- Tissue processing
- Turn the container upside down and allow air between the agar and the sides of the container to remove the solid agar solution, including the heart, from the container. Remove the plastic rods from the solid agar.
- Section the agar block containing the heart in 5-mm slices from the apex to the base of the heart using a meat slicer. Keep the angulation of the cut slices the same as in the acquired MR images by cutting parallel to the bottom of the agar block.
- Stain the agar slices (including the heart) for 15 min in 1 wt% of 2,3,5-triphenyltetrazoliumchloride (TTC) dissolved in 0.9% saline at 37 °C, and photograph the slices on both sides from a perpendicular view (Figure 2A). Next, carefully rinse the slices in 0.9% saline.
NOTE: In this study, we used a dSLR setup with an appropriate lens/objective, a tripod, and uniform lighting. However, the photographs served only as a control for the assessment of the scar region, so we could have used a different setup.
- Fluorescence imaging
NOTE: Depending on the excitation and emission wavelengths of the fluorescent microbeads, select the appropriate filter block and excitation lasers (e.g., the red microbeads used here have excitation and emission wavelengths of 580 nm and 605 nm, respectively; therefore, the selected excitation laser and bandpass filters were set to 532 nm, 580/30 nm and 610/30 nm, respectively).- Select fluorescence-mode imaging on the variable-mode scanner. Set the photomultiplier tube to 430 V or equivalent and the pixel size to 100 x 100 µm. Select an excitation laser (532 nm) closest to the excitation wavelength of the fluorescent microbeads.
- For the first filter block, select a bandpass filter (580/30 nm) that overlaps with the emission wavelength of the injected fluorescence beads (channel 1). Select a bandpass filter for the second filter block (610/30) outside the emission wavelength (channel 2).
NOTE: The second filter block serves as a negative control and to remove auto-fluorescence while keeping the injection sites intact. - Scan both sides of the agar slices in the fluorescence mode of the variable-mode laser scanner using the two channels. Make sure that each slice is completely scanned, including the reference holes.
5. Post-processing
NOTE: The first step in image post-processing is the manual segmentation of the myocardium using in-house developed scripts to trace the endo- and epicardial borders, as well as the injection sites. This is the same for both MRI and fluorescence scans.
- Segment the myocardium in the MRI scans.
- Segment the endocardial and epicardial LV borders on the FLAIR MRI sequence images.
- Copy the LV segmentation from step 5.1.1 to the LGE-MRI dataset and segment the scar on the LGE MRI sequence.
- Copy the myocardium segmentation from step 5.1.1 to the T2*-weighted dataset and segment the SPIO depositions in the LV myocardium.
- Process the fluorescence images and perform segmentations.
- Load the files obtained from the variable-mode scanner and make a separate image of each cross-sectional heart slice.
- Flip the slices that were scanned in base to apex orientation and sort the fluorescence images into a stack for both channels that is oriented from apex to base.
- Segment the endocardial and epicardial LV borders on the fluorescence images.
- Segment the scar manually on the fluorescence images and use the LGE-MRI scan and the photographs to confirm scar morphology.
- Subtract the image stack of channel 2 from the image stack of channel 1 to exclude auto-fluorescence. Manually segment the fluorescent microbead depositions and use the T2* images for confirmation.
- To create an anatomically-correct 3D geometry, perform a rigid registration of the slices in the image stack based on the reference structures (the holes created by the rigid tubes). Calculate and store the applied translation and rotation of each image.
- Apply the stored transformations to the image stacks and the segmentations. Linearly interpolate the segmentations of both sides of the slices to reconstruct the original slice thickness and to create a 3D model of the data.
6. Analysis
- Perform 2D and/or 3D measurements of the distance between the centers of the injection sites and the IBZ to assess the injection accuracy. Measure the distance along the endocardial border of the LV segmentation. In Figure 2C and 2F, an example of the 2D and 3D measurements is indicated by the red line.
Wyniki
Tissue Embedding
Through the embedding process, an end-diastolic-like geometry was established. The agar successfully adhered to the heart tissue, enabling the tissue to be sliced at the desired angulation with equal slice thicknesses (Figure 2A and 2C).
Scar- and Injection-site Assessment
Dyskusje
Whole-heart 3D myocardial tissue processing according to this protocol provides a structured method that enables the 3D analysis of the infarct, the IBZ, and the performed injections with respect to the cardiac anatomy. The filling volume of the heart depends on the desired analysis. In this study, to assess the injection accuracy, we aimed to fill the heart to resemble the end-diastolic geometry as closely as possible. To enforce this, the LV apex is fixed to the bottom of the container and the LV is filled with agar wh...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
The authors would like to thank Marlijn Jansen, Joyce Visser, and Martijn van Nieuwburg for their assistance with the animal experiments. We greatly acknowledge Martijn Froeling and Anke Wassink for their assistance with the MRI imaging.
Materiały
| Name | Company | Catalog Number | Comments |
| 0.9% Saline | Braun | ||
| Agarose | Roche Diagnostics | Scientific grade multipurpose agar | |
| Biomolecular fluorescence scanner Typhoon 9410 | GE Healthcare | ||
| Embedding container | Plastic, dimensions 17 x 14.5 x 14 cm | ||
| FluoSpheres Polystyrene Microspheres | Invitrogen | F8834 | red, 10 µm |
| Gadolinium | Gadovist | 1.0 mmol/mL | |
| dS 32 channel head coil | Philips | Or similar | |
| Matlab | Mathworks | To insure compatability 2015a or newer | |
| Meat slicer | Berkel | ||
| Myostar injection catheter | Biosense Webster | ||
| Super paramagnetic iron oxide particles | Sinerem | ||
| Triphenyl-tetrazolium chloride | Merck | ||
| UPy-PEG10k | |||
| Vicryl 2-0 | Ethicon |
Odniesienia
- Nowbar, A. N., Howard, J. P., Ja Finegold, ., Asaria, P., Francis, D. P. 2014 global geographic analysis of mortality from ischaemic heart disease by country, age and income: statistics from World Health Organisation and United Nations. Int J Cardiol. 174 (2), 293-298 (2014).
- Kannel, W. B., Belanger, A. J. Epidemiology of heart failure. Am Heart J. 121 (3), 951-957 (1991).
- Ibáñez, B., Heusch, G., Ovize, M., Van De Werf, F. Evolving therapies for myocardial ischemia/reperfusion injury. J Am Coll Cardiol. 65 (14), 1454-1471 (2015).
- Bartunek, J., Vanderheyden, M., Hill, J., Terzic, A. Cells as biologics for cardiac repair in ischaemic heart failure. Heart. 96 (10), 792-800 (2010).
- Orlic, D., Kajstura, J., et al. Bone marrow cells regenerate infarcted myocardium. Nature. 410 (6829), 701-705 (2001).
- Nguyen, P. K., Lan, F., Wang, Y., Wu, J. C. Imaging: Guiding the Clinical Translation of Cardiac Stem Cell Therapy. Circ Res. 109 (8), 962-979 (2011).
- Psaltis, P. J., Worthley, S. G. Endoventricular electromechanical mapping-the diagnostic and therapeutic utility of the NOGA XP Cardiac Navigation System. J Cardiovasc Transl Res. 2 (1), 48-62 (2009).
- Tomkowiak, M. T., Klein, A. J., et al. Targeted transendocardial therapeutic delivery guided by MRI-x-ray image fusion. Catheter Cardiovasc Interv. 78 (3), 468-478 (2011).
- Dauwe, D. F., Nuyens, D., et al. Three-dimensional rotational angiography fused with multimodal imaging modalities for targeted endomyocardial injections in the ischaemic heart. Eur Heart J Cardiovasc Imaging. 15 (8), 900-907 (2014).
- van Slochteren, F. J., van Es, R., et al. Multimodality infarct identification for optimal image-guided intramyocardial cell injections. Neth Heart J. 22 (11), 493-500 (2014).
- van Slochteren, F. J., van Es, R., et al. Three dimensional fusion of electromechanical mapping and magnetic resonance imaging for real-time navigation of intramyocardial cell injections in a porcine model of chronic myocardial infarction. Int J Cardiovasc Imaging. 32 (5), 833-843 (2016).
- Pape, a. C. H., Bakker, M. H., et al. An Injectable and Drug-loaded Supramolecular Hydrogel for Local Catheter Injection into the Pig Heart. J Vis Exp. (100), (2015).
- Bastings, M. M. C., Koudstaal, S., et al. A fast pH-switchable and self-healing supramolecular hydrogel carrier for guided, local catheter injection in the infarcted myocardium. Adv Healthc Mater. 3 (1), 70-78 (2014).
- Koudstaal, S., Jansen of Lorkeers, S. J., et al. Myocardial infarction and functional outcome assessment in pigs. J. Vis. Exp. (86), (2014).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone