Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
TACI: An ImageJ Plugin for 3D Calcium Imaging Analysis
W tym Artykule
Podsumowanie
TrackMate Analysis of Calcium Imaging (TACI) is an open-source ImageJ plugin for 3D calcium imaging analysis that examines motion on the z-axis and identifies the maximum value of each z-stack to represent a cell's intensity at the corresponding time point. It can separate neurons overlapping in the lateral (x/y) direction but on different z-planes.
Streszczenie
Research in neuroscience has evolved to use complex imaging and computational tools to extract comprehensive information from data sets. Calcium imaging is a widely used technique that requires sophisticated software to obtain reliable results, but many laboratories struggle to adopt computational methods when updating protocols to meet modern standards. Difficulties arise due to a lack of programming knowledge and paywalls for software. In addition, cells of interest display movements in all directions during calcium imaging. Many approaches have been developed to correct the motion in the lateral (x/y) direction.
This paper describes a workflow using a new ImageJ plugin, TrackMate Analysis of Calcium Imaging (TACI), to examine motion on the z-axis in 3D calcium imaging. This software identifies the maximum fluorescence value from all the z-positions a neuron appears in and uses it to represent the neuron's intensity at the corresponding t-position. Therefore, this tool can separate neurons overlapping in the lateral (x/y) direction but appearing on distinct z-planes. As an ImageJ plugin, TACI is a user-friendly, open-source computational tool for 3D calcium imaging analysis. We validated this workflow using fly larval thermosensitive neurons that displayed movements in all directions during temperature fluctuation and a 3D calcium imaging dataset acquired from the fly brain.
Wprowadzenie
The level of intracellular calcium is a precise marker of neuronal excitability. Calcium imaging measures the changes in intracellular calcium to understand neuronal activity1. Studies in neuroscience have increasingly used this method due to the development of techniques for measuring intracellular calcium concentration, including genetically encoded calcium indicators (GECIs), such as GCaMP2,3, which can be noninvasively expressed in specific sets of neurons through genetic approaches. The lower costs of lasers and microscope components have also increased the use of calcium imaging4. Importantly, calcium imaging allows for recording and studying single neurons as well as large neuron populations simultaneously in freely moving animals5.
Nevertheless, the analysis of calcium imaging data is challenging because (1) it involves tracking the changes in fluorescence of individual cells over time, (2) the fluorescence signal intermittently disappears or reappears with neuronal responses, and (3) the neurons may move in all directions, specifically in and out of a focal plane or appearing on multiple planes4,6. Manual analysis is time-consuming and becomes impractical as the length of recordings and the number of neurons increases. Various software programs have been developed to accelerate the process of analyzing calcium imaging. Previously, software was designed in a limited experimental context, making it difficult for other laboratories to adopt it. Recent efforts to meet modern standards for software sharing have led to the development of several tools that can consistently analyze calcium imaging data across different groups7,8,9,10,11,12,13,14,15,16,17,18,19. However, most of these tools require programming knowledge and/or depend on commercial software. A lack of programming knowledge and software paywalls deter researchers from adopting these methods. Moreover, many of these tools focus on correcting the x/y motion, although motion on the z-axis also needs to be explicitly diagnosed and corrected6. There is a need for a computational tool to analyze 3D calcium imaging that focuses on neurons exhibiting z-drift and appearing on multiple z-planes. Ideally, this tool should use open-source software and not require programming knowledge to allow other laboratories to readily adopt it.
Here, we developed a new ImageJ plugin, TACI, to analyze 3D calcium imaging data. First, the software renames, if needed, and organizes the 3D calcium imaging data by z-positions. The cells of interest are tracked in each z-position, and their fluorescence intensities are extracted by TrackMate or other computational tools. TACI is then applied to examine the motion on the z-axis. It identifies the maximum value of a z-stack and uses it to represent a cell's intensity at the corresponding time point. This workflow is suited to analyzing 3D calcium imaging with motion in all directions and/or with neurons overlapping in the lateral (x/y) direction but appearing in different z-positions. To validate this workflow, 3D calcium imaging datasets from fly larval thermosensitive neurons and mushroom neurons in the brain were used. Of note, TACI is an open-source ImageJ plugin and does not require any programming knowledge.
Access restricted. Please log in or start a trial to view this content.
Protokół
1. Calcium imaging
- Fly larvae preparation
NOTE: Flies and larvae are maintained at 25 °C under a 12 h:12 h light:dark cycle.- Anesthetize the flies with CO2. Sort 20-45 males and 20-45 females into each fly vial, and give them at least 24 h to 48 h to recover from the CO2 exposure.
NOTE: Fly exposure to CO2 should last for the shortest amount of time possible. - To synchronize the larvae age, tap over the flies into new vials containing yeast granules, and allow them 4-8 h to lay eggs. Remove the flies by flipping them into new vials.
- Collect the larvae at 72 h using 10 mL of 20% w/v sucrose solution.
- Anesthetize the flies with CO2. Sort 20-45 males and 20-45 females into each fly vial, and give them at least 24 h to 48 h to recover from the CO2 exposure.
- Microscope and temperature control setup
- Perform the imaging on a confocal microscope (see the Table of Materials) and a z-axis piezo stage with a stage insert using the following settings: laser, Argon; scan mode, frame; frame size, 512 x 512; speed, maximum; channels/bit depth, 1/8 Bit; zoom, 1.5; z-stack, slice =15, Keep = slice; focus devices and strategy, Definite focus; focus, Definite focus on.
- Attach a Peltier cooling module to a heat sink by the heat transfer compound to build a thermoelectric cooler. The Peltier is powered by a 2 A power supply.
- Attach a thermocouple microprobe to a data acquisition device to record the temperature.
- Calcium imaging
- Rinse the larvae 3x in 1x phosphate-buffered saline (PBS).
- Pipette 75 μL of 1x PBS onto the center of a glass slide.
- Put one or two larvae in 1x PBS and place the thermocouple microprobe near the larvae.
NOTE: The distance between the larvae and the microprobe should be ~5 mm. The larvae may move if the microprobe is positioned too close to them. A great distance could result in inaccurate temperature readings. - Cover the larvae and thermocouple microprobe with a glass coverslip. Seal the coverslip with nail polish.
- Place the slide on the microscope stage, find the focus using a 25x objective, and place the thermoelectric cooler on the slide.
NOTE: The Peltier is placed directly on the slide where the larvae are to deliver the temperature stimuli. - In confocal software, focus on the fluorescent cells of interest. Adjust the laser power to avoid oversaturation. Set the first and last slice positions in the z-stack setting.
NOTE: Use the lowest possible laser power before recording to prevent photobleaching. - Start the z-stack scanning and temperature recording at the same time.
- Control the power supply that powers the Peltier to change the Peltier surface temperature. Turn on the power supply to decrease the temperature and turn it off to increase the temperature.
- Stop the z-stack scanning and temperature recording.
2. Analysis of the 3D calcium imaging data
- Export the calcium imaging data to TIFF files and save them in a folder with the same name as the base name of the TIFF files inside.
NOTE: The filename must not contain any commas.- Use the following parameters to export the calcium imaging data from ZEN (black edition): file type, TIFF; compression, LZW; channels, 2; Z-position, all; time, all; phase, 1; region, full.
- Installation
- Download TACI-Calcium_Imaging.jar from Github (https://github.com/niflylab/TACI_CalciumImagingPlugin/releases).
- Install the plugin in FIJI by clicking on Plugins in the menu bar and then clicking on Install in the dropdown menu (Plugins | Install). Then, restart FIJI.
NOTE: Do NOT use Plugins | Install Plugin. - Run the plugin by clicking on Plugins and then choosing TACI-Calcium Imaging (Plugins | TACI-Calcium Imaging).
- Use the RENAME function to convert the TIFF filenames to the required structure.
NOTE: The tool uses filename_h#t#z#c#.tif as the default structure (h# and c# are optional; #: a positive integer). If the image filenames are not in the default structure, the RENAME function must be executed.- Choose the folder in which the TIFF images need renaming by clicking Browse Folders.
- Fill in the parameter information. Five parameters are listed, including Filename, Phase, Max T-Position, Max Z-Position, and Channel. Each parameter has three values: Preceding Text, Parameter Value, and Order.
NOTE: The information is case-sensitive. If the image filenames contain a phrase before a parameter, fill in the phrase as the Preceding Text of the corresponding parameter. If the image filenames contain a phrase after all parameters, fill in the phrase as the Post Text.- Be sure to enter Filename, Max T-Position, and Max Z-Position and that the parameter values for Max T-Position and Max Z-Position include all digits.
- Wait for the Filename to be automatically filled using the folder name.
- If the TIFF filenames do not include the phase and channel, leave the corresponding parameter value blank, and choose Na in Order.
- Click Rename to create a folder with the same name and _r. Observe that in the folder, the TIFF filenames have been restructured to be compatible with the ORGANIZE function.
NOTE: _r has been added to the filenames of the TIFF images.
- Use the ORGANIZE function to save the TIFF images from the same z-position in one folder.
NOTE: The folder name must have the same name as the base name of the TIFF files inside and must NOT have any commas.- Choose the folder in which the TIFF images need organizing by clicking Browse Folders.
- If the parameter CSV file (param.csv) exists, wait for the parameter values to be filled in automatically.
NOTE: The param.csv file has a required format. The parameters, including filename, phase, position_t, position_z, channel, and is_gray, must be filled in in row 1 from left to right starting from column A. Corresponding values for each parameter must be filled in in row 2. - If the parameter CSV file (param.csv) does not exist, manually fill in the parameter values. Ensure that the parameter values of Phase and Channel include letters, while the parameter values of T Position and Z Position should be the largest numbers of the t- and z-positions. If the image filenames do not include the phase or channel, enter Na.
- Create grayscale TIFF images when needed. Leave the box of Are images gray? unchecked to grayscale the images.
- Click Organize to create a folder with the same name and _gray_stacks and to generate folders with the same name and _# (#: z-positions) in the folder. Observe that the TIFF files are sorted into corresponding folders by z-positions and that a file named param.csv is generated, in which the parameters and their values can be found.
- Use TrackMate in FIJI to extract the fluorescence intensities of the cells of interest from each z-position.
NOTE: This step can also be accomplished by other imaging software.- Open the TIFF images in a z-position folder by FIJI.
- Run TrackMate by clicking on Plugins | Tracking | TrackMate, and adjust the following parameters if necessary.
- Use DoG or LoG detectors.
- Change the blob diameter, threshold, and median filter. Adjust the blob diameter to be similar to the diameter of the cells. If the cells are oval, adjust the blob diameter to be similar to the minor axis.
NOTE: Increasing the threshold helps avoid background noise being picked up as signals without affecting the signal intensities (Supplementary Figure 1). However, an increase in the threshold may miss true signals (Supplementary Figure 1). If the signals are strong, use the median filter to decrease the Salt and Pepper noise. - Set the filters to remove some, if not all, of the irrelevant signals. Filters X and Y are spatial filters. Use filters X and Y to remove the irrelevant signals that are distant from the real signals.
NOTE: When filters are set on one image, it is crucial to check all the other images to ensure that the real signals are not removed. - Set the linking max distance, gap-closing max distance, and gap-closing max frame gap. Set the linking max distance and gap-closing max distance to be 3x-5x the blob diameter, especially when the samples move significantly over time, to help decrease the number of tracks. Set the gap-closing max frame gap to the number of images in the stack.
- Export the fluorescence intensities of the regions of interest (ROIs) to a CSV file. If an old TrackMate version is used, choose Export all spots statistics in the Select an action window. If the TrackMate version is 7.6.1 or higher, choose the Spots in the Display options window. Export or save as the interactive files to CSV files.
NOTE: Both files are interactive with the image window; highlighting an ROI displays the corresponding ROI in the image window. These files include the mean intensities (MEAN_INTENSITY or MEAN_INTENSITY_CH1) of the cells of interest at corresponding time points (POSITION_T). The same TRACK_IDs should represent the same ROIs at different time points. However, this is not always true and may need to be corrected manually, when necessary. If TrackMate does not recognize the ROIs at some time points, those time points are not displayed.
- Extract the background fluorescence intensity, and create the Background_list.csv file.
NOTE: In this study, the background intensity for each z-position was estimated by using the average value of three to five neighboring same-size blobs that did not contain fluorescence signals and were from different time points.- The Background_list.csv file has a required format: each column contains the information of one neuron, starting from Neuron 0. Fill in the neuron numbers, such as Neuron 0, in row 1. Then, provide the background intensity for each z-position analyzed-if five z-positions are analyzed for Neuron 0, fill in five background intensity values below Neuron 0.
NOTE: The Background_list.csv file is required for the TACI EXTRACT function. If the background is negligible, the Background_list.csv with the zero-background intensity of every neuron must be provided.
- The Background_list.csv file has a required format: each column contains the information of one neuron, starting from Neuron 0. Fill in the neuron numbers, such as Neuron 0, in row 1. Then, provide the background intensity for each z-position analyzed-if five z-positions are analyzed for Neuron 0, fill in five background intensity values below Neuron 0.
- Use the EXTRACT function to identify the maximum fluorescence intensities at each t-position and calculate ΔF/F0 as shown in equation (1).
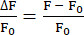 (1)
(1)
- Create a folder and name it using the neuron number, starting from Neuron 0. Save the CSV files containing the fluorescence information of the corresponding neuron in the folder. Each CSV file contains the information of one z-position, so the number of CSV files equals the number of z-positions analyzed. Name the CSV files as Mean_Intensity#.csv (# represents the z-position) and each CSV file includes at least two columns: POSITION_T and MEAN_INTENSITY or MEAN_INTENSITY_CH1.
NOTE: Create a folder for every cell of interest. - Save the Background_list.csv file and folders created in step 2.7.1 in one folder. Choose this folder by clicking on Browse Files.
NOTE: The number of background values in the Background_list.csv file must match the number of the Mean_Intensity#.csv files for each neuron. - The Background File is automatically filled in. Fill in the largest number of t-positions for the Number of T Positions.
- Click Extract to create a results folder, including the CSV files and plots for each neuron. The CSV files include information on the maximum fluorescence intensity and ΔF/F0 at each t-position. The plots are line charts of ΔF/F0 over t-positions.
NOTE: In this study, ΔF/F0 was calculated using equation (1). The first value of each z-position was used as F0. If this F0 is not appropriate20, the plugin provides files including raw data for each neuron in the python_files folder.
- Create a folder and name it using the neuron number, starting from Neuron 0. Save the CSV files containing the fluorescence information of the corresponding neuron in the folder. Each CSV file contains the information of one z-position, so the number of CSV files equals the number of z-positions analyzed. Name the CSV files as Mean_Intensity#.csv (# represents the z-position) and each CSV file includes at least two columns: POSITION_T and MEAN_INTENSITY or MEAN_INTENSITY_CH1.
- Use the MERGE function to average each neuron's ΔF/F0, calculate the SEM, and plot the average ΔF/F0 over t-positions.
- Choose the results folder created by the EXTRACT function by clicking on Browse Files.
- Fill in the Number of T Positions with the largest number of t-positions.
- Click Merge to create a merged_data folder, including a merged_data.csv file and an Average_dF_F0.png plot. The CSV file includes the information on the average and SEM ΔF/F0 at each t-position. The plot is a line chart of the average ΔF/F0 over t-positions.
Access restricted. Please log in or start a trial to view this content.
Wyniki
Workflow of 3D calcium imaging analysis
In this study, we developed a new ImageJ plugin, TACI, and described a workflow to track z-drift and analyze 3D calcium imaging that pinpoints the responses of individual cells appearing in multiple z-positions (Figure 1). This tool has four functions: RENAME, ORGANIZE, EXTRACT, and MERGE. First, if the image names are not compatible with the ORGANIZ...
Access restricted. Please log in or start a trial to view this content.
Dyskusje
This study developed a new ImageJ plugin, TACI, and described a workflow analyzing 3D calcium imaging. Many currently available tools focus on correcting the x/y motion, although motion on the z-axis also needs to be explicitly diagnosed or corrected6. During image acquisition in a live organism, movement on the z-axis is unavoidable even when the organism is immobilized, and some stimuli, such as temperature change, often cause significant z-drift. Increasing the height of the z-stacks will allow...
Access restricted. Please log in or start a trial to view this content.
Ujawnienia
The authors have no conflicts of interest to disclose.
Podziękowania
A Zeiss LSM 880 in the Fralin Imaging Center was used to collect the calcium imaging data. We acknowledge Dr. Michelle L Olsen and Yuhang Pan for their assistance with the IMARIS software. We acknowledge Dr. Lenwood S. Heath for constructive comments on the manuscript and Steven Giavasis for comments on the GitHub README file. This work was supported by NIH R21MH122987 (https://www.nimh.nih.gov/index.shtml) and NIH R01GM140130 (https://www.nigms.nih.gov/) to L.N. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Access restricted. Please log in or start a trial to view this content.
Materiały
| Name | Company | Catalog Number | Comments |
| Blunt Fill Needel | BD | 303129 | |
| Calcium chloride dihydrate | Fisher Scientific | 10035-04-8 | Fly food ingredient |
| Carbon dioxide | Airgas | UN1013 | Size 200 High Pressure Steel Cylinder |
| CO2 bubbler kit | Genesee | 59-180 | |
| Confocal microscope LSM880 | Zeiss | 4109002107876000 | An inverted Axio Observer Z1, equipped with 5 lasers, 2 standard PMT detectors, 32-channel GaAsP dectectors, an Airyscan detector, and Definite Focus.2. |
| DAQami software | Measurement Computing | ||
| Dextrose | Genesee | 62-113 | Fly food ingredient |
| Drosophila Agar | Genesee | 66-111 | Fly food ingredient |
| Ethanol | Decon Labs, Inc. | 64-17-5 | Fly food ingredient |
| Fly line: Ir21a-Gal4 | Dr. Paul Garrity lab | A kind gift | |
| Fly line: Ir21a-Gal80 | Dr. Lina Ni lab | ||
| Fly line: Ir68a-Gal4 | Dr. Aravinthan DT Samuel lab | A kind gift | |
| Fly line: Ir93a-Gal4 | Dr. Paul Garrity lab | A kind gift | |
| Fly line: UAS-GCaMP6 | Bloomington Drosophila Stock Center | 42750 | |
| Flypad | Genesee | 59-114 | |
| General purpose forged brass regulator | Gentec | G152 | |
| Gibco PBS pH 7.4 (1x) | Thermo Fisher Scientific | 10010-031 | |
| Green Drosophila tubing | Genesee | 59-124 | |
| Heat transfer compound | MG Chemicals | 860-60G | |
| Heatsink | Digi-Key Electronics | ATS2193-ND | Resize to 12.9 x 5.5 cm |
| Illuminator | AmScope | LED-6W | |
| Inactive Dry Yeast | Genesee | 62-108 | Fly food ingredient |
| Incubator | Pervical | DR-41VL | Light: dark cycle: 12h:12h; temperature: 25 °C; humidity: 40-50% RH. |
| Methyl-4-hydroxybenzoate | Thermo Scientific | 126965000 | Fly food ingrediete |
| Micro cover glass | VWR | 48382-126 | 22 x 40 mm |
| Microscope slides | Fisher Scientific | 12-544-2 | 25 x 75 x 1.0 mm |
| Nail polish | Kleancolor | ||
| Narrow Drosophila vials | Genesee | 32-113RL | |
| Objective | Zeiss | 420852-9871-000 | LD LCI Plan-Apochromat 25x/0.8 Imm Corr DIC M27 |
| Peltier cooling module | TE Technology | TE-127-1.0-0.8 | 30 x 30 mm |
| Plugs | Genesee | 49-102 | |
| Power Supply | Circuit Specialists | CSI1802X | 10 volt DC 2.0 amp linear bench power supply |
| Princeton Artist Brush Nepture | Princeton Artist Brush Co. | Series 4750, size 2 | |
| Sodium potassium L-tartrate tetrahydrate | Thermo Scientific | 033241-36 | Fly food ingredient |
| Stage insert | Wienecke and Sinske | 432339-9030-000 | |
| Stereo Microscope | Olympus | SZ61 | Any stereo microscope works |
| T-Fitting | Genesee | 59-123 | |
| Thermocouple data acquisition device | Measurement Computing | USB-2001-TC | Single channel |
| Thermocouple microprobe | Physitemp | IT-24P | |
| Yellow Cornmeal | Genesee | 62-101 | Fly food ingredient |
| Z-axis piezo stage | Wienecke and Sinske | 432339-9000-000 |
Odniesienia
- Grienberger, C., Konnerth, A. Imaging calcium in neurons. Neuron. 73 (5), 862-885 (2012).
- Nakai, J., Ohkura, M., Imoto, K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nature Biotechnology. 19 (2), 137-141 (2001).
- Zhang, Y., et al. jGCaMP8 fast genetically encoded calcium indicators. Janelia Research Campus. , (2020).
- Robbins, M., Christensen, C. N., Kaminski, C. F., Zlatic, M. Calcium imaging analysis - How far have we come. F1000Research. 10, 258(2021).
- Oh, J., Lee, C., Kaang, B. K. Imaging and analysis of genetically encoded calcium indicators linking neural circuits and behaviors. The Korean Journal of Physiology & Pharmacology. 23 (4), 237-249 (2019).
- Stringer, C., Pachitariu, M. Computational processing of neural recordings from calcium imaging data. Current Opinion in Neurobiology. 55, 22-31 (2019).
- Pnevmatikakis, E. A., Giovannucci, A. NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. Journal of Neuroscience Methods. 291, 83-94 (2017).
- Nguyen, J. P., Linder, A. N., Plummer, G. S., Shaevitz, J. W., Leifer, A. M. Automatically tracking neurons in a moving and deforming brain. PLoS Computational Biology. 13 (5), 1005517(2017).
- Lagache, T., Hanson, A., Pérez-Ortega, J. E., Fairhall, A., Yuste, R. EMC2: A versatile algorithm for robust tracking of calcium dynamics from individual neurons in behaving animals. bioRxiv. , (2021).
- Giovannucci, A., et al. CaImAn an open source tool for scalable calcium imaging data analysis. Elife. 8, 38173(2019).
- Delestro, F., et al. In vivo large-scale analysis of Drosophila neuronal calcium traces by automated tracking of single somata. Scientific Reports. 10, 7153(2020).
- Cantu, D. A., et al. EZcalcium: Open-source toolbox for analysis of calcium imaging data. Frontiers in Neural Circuits. 14, 25(2020).
- Eglen, S. J., et al. Toward standard practices for sharing computer code and programs in neuroscience. Nature Neuroscience. 20 (6), 770-773 (2017).
- Pachitariu, M., et al. Suite2p: Beyond 10,000 neurons with standard two-photon microscopy. bioRxiv. , (2017).
- Corder, G., et al. An amygdalar neural ensemble that encodes the unpleasantness of pain. Science. 363 (6424), 276-281 (2019).
- Lagache, T., Hanson, A., Pérez-Ortega, J. E., Fairhall, A., Yuste, R. Tracking calcium dynamics from individual neurons in behaving animals. PLoS Computational Biology. 17 (10), 1009432(2021).
- Kolar, K., Dondorp, D., Zwiggelaar, J. C., Høyer, J., Chatzigeorgiou, M. Mesmerize is a dynamically adaptable user-friendly analysis platform for 2D and 3D calcium imaging data. Nature Communications. 12, 6569(2021).
- Moein, M., et al. CaSiAn: A Calcium Signaling Analyzer tool. Bioinformatics. 34 (17), 3052-3054 (2018).
- Zhou, P., et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Elife. 7, 28728(2018).
- Neugornet, A., O'Donovan, B., Ortinski, P. I. Comparative effects of event detection methods on the analysis and interpretation of Ca(2+) imaging data. Frontiers in Neuroscience. 15, 620869(2021).
- Tinevez, J. Y., et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2017).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Fazeli, E., et al. Automated cell tracking using StarDist and TrackMate. F1000Research. 9, 1279(2020).
- Chen, T. W., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 499 (7458), 295-300 (2013).
- Ni, L., et al. The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. Elife. 5, 13254(2016).
- Omelchenko, A. A., et al. Cool and warm ionotropic receptors control multiple thermotaxes in Drosophila larvae. Frontiers in Molecular Neuroscience. , (2022).
- Sanchez-Alcaniz, J. A., et al. An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nature Communications. 9 (1), 4252(2018).
- Hernandez-Nunez, L., et al. Synchronous and opponent thermosensors use flexible cross-inhibition to orchestrate thermal homeostasis. Science Advances. 7 (35), (2021).
- Klein, M., et al. Sensory determinants of behavioral dynamics in Drosophila thermotaxis. Proceedings of the National Academy of Sciences of the United States of America. 112 (2), 220-229 (2015).
Access restricted. Please log in or start a trial to view this content.
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone