A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Evaluation of the Spatial Distribution of γH2AX following Ionizing Radiation
In This Article
Summary
Microscopic analysis of γH2AX foci, which form following the phosphorylation of H2AX at Ser-139 in response to DNA double-strand breaks, has become an invaluable tool in radiation biology. Here we used an antibody to mono-methylated histone H3 at lysine 4 as an epigenetic marker of actively transcribing euchromatin, to evaluate the spatial distribution of radiation-induced γH2AX formation within the nucleus.
Abstract
An early molecular response to DNA double-strand breaks (DSBs) is phosphorylation of the Ser-139 residue within the terminal SQEY motif of the histone H2AX1,2. This phosphorylation of H2AX is mediated by the phosphatidyl-inosito 3-kinase (PI3K) family of proteins, ataxia telangiectasia mutated (ATM), DNA-protein kinase catalytic subunit and ATM and RAD3-related (ATR)3. The phosphorylated form of H2AX, referred to as γH2AX, spreads to adjacent regions of chromatin from the site of the DSB, forming discrete foci, which are easily visualized by immunofluorecence microscopy3. Analysis and quantitation of γH2AX foci has been widely used to evaluate DSB formation and repair, particularly in response to ionizing radiation and for evaluating the efficacy of various radiation modifying compounds and cytotoxic compounds4.
Given the exquisite specificity and sensitivity of this de novo marker of DSBs, it has provided new insights into the processes of DNA damage and repair in the context of chromatin. For example, in radiation biology the central paradigm is that the nuclear DNA is the critical target with respect to radiation sensitivity. Indeed, the general consensus in the field has largely been to view chromatin as a homogeneous template for DNA damage and repair. However, with the use of γH2AX as molecular marker of DSBs, a disparity in γ-irradiation-induced γH2AX foci formation in euchromatin and heterochromatin has been observed5-7. Recently, we used a panel of antibodies to either mono-, di- or tri- methylated histone H3 at lysine 9 (H3K9me1, H3K9me2, H3K9me3) which are epigenetic imprints of constitutive heterochromatin and transcriptional silencing and lysine 4 (H3K4me1, H3K4me2, H3K4me3), which are tightly correlated actively transcribing euchromatic regions, to investigate the spatial distribution of γH2AX following ionizing radiation8. In accordance with the prevailing ideas regarding chromatin biology, our findings indicated a close correlation between γH2AX formation and active transcription9. Here we demonstrate our immunofluorescence method for detection and quantitation of γH2AX foci in non-adherent cells, with a particular focus on co-localization with other epigenetic markers, image analysis and 3D-modeling.
Protocol
Cell preparation
- Human erythroleukemic K562 cells are grown in RPMI-1640 medium supplemented with 10% (v/v) fetal bovine serum and 20 mg/ml gentamicin in a humidified 5% CO2 environment at 37°C.
- Approximately 18 hours prior to staining wash exponentially growing cells (optimal at 5 x 105 cells/ml) with phosphate buffered saline (PBS), resuspend in fresh media and return to 37°C, 5% CO2.
- Wash the cells twice with PBS by centrifugation at approximately 1500 rpm for 5 minutes and resuspend in fresh media.
- Count cells and adjust the cell density to approximately 5 x 105 cells/ml.
Cells can be counted either by an automated method (e.g. Sysmex or Coulter counter with filter size between 5 to 20 microns) or by using the trypan blue exclusion method and a haemocytometer. Excess number of cells (>800 cells/cm2) may lead to non-uniform staining.
Irradiation and immunofluorescence staining
- Assemble cytospin clips, filter cards and slides with appropriate labelling.
When assembling the cytospin apparatus ensure that the hole in filter card coincides with the funnel.
- Expose cells to either γ-radiation or X-rays (2Gy in this example).
To prevent the formation of foci during the irradiation, keep the cells on ice for 5 to 10 minutes prior to and during irradiation.
Following irradiation incubate the cells at 37°C, 5% CO2 for the required time (For peak γH2AX levels 30 minutes to 1 hour; 1 hour in this example).
- Wash the cells twice with cold PBS by centrifugation and resuspend in fresh PBS.
- Dispense 100 to 150 μL of the cell suspension into each cytospin funnel and spin at 500 rpm for 5 minutes.
- Separate the slides from cytospins and allow the cells to moderately air dry for approximately 15 minutes.
Do not let cells dry completely as cellular morphology may disintegrate.
- Draw a circle around the cells with a hydrophobic pen.
- Fix cells with 4% paraformaldehyde for 5 minutes at room temperature (RT).
In our experience, fixation with paraformaldehyde is superior to fixing with ethanol or methanol. Suspension cells require fixation for 5 minutes at RT whereas longer fixation times (15 to 20 minutes) are needed for optimal fixation of adherent cells.
We find that staining immediately yields better results than storing the fixed slides at -20°C which typically results in high background fluorescence.
- Wash the cells twice with PBS for 5 minutes each.
- Permeabilize the cells with 0.1% Triton-X 100 at RT for 5 minutes.
Compared to Tween-80 we typically observe better signals in cells permeabilized with Triton-X 100. Depending on cell type it may be necessary to change the permeabilisation time. For example, in K562 cells permeabilization for 5 minutes at RT is sufficient whereas in adherent cells a 15 minute permeabilization is required.
- Wash cells three times with PBS for 5 minutes each.
- Block cells in 1% BSA for 30 minute (3 x 10 minutes) at RT.
We compared the blocking efficiency using serum (derived from same species as the secondary antibodies) and bovine serum albumin in PBS. BSA was more efficient in minimizing non-specific binding of secondary antibodies compared to serum. We compared different blocking times, overnight blocking at 4°C compared to 3 x 10 minute and 3 x 20 minute blocking steps at room temperature, and determined that blocking for 3 x 10 minutes was optimal.
- Add 50 μL of primary antibody (1: 500 diluted in 1% BSA) to each slide.
Incubation with primary antibody at RT for 60 minutes results in reduced background staining compared to an overnight incubation at 4°C.
- To stain different markers simultaneously use antibodies raised in different species (e.g. mouse anti-γH2AX and rabbit anti-methylated H3K4 in this example).
- Incubate with primary antibodies for 90 minutes at RT on a rocking platform at 250 rpm.
All incubations are performed in a humidified staining trough.
- Wash cells three times with PBS for 5 minutes each.
- Add 50 μL of (1: 1000 diluted in 1% BSA) secondary antibody to each slide.
To stain for different markers on the same slide use secondary antibodies with different species specificity (according to the primary antibody) and fluorescence emission wavelength (e.g. anti-mouse Alexa 488 (green) and anti-rabbit Alexa-546 (red) in this example).
- Incubate cells for 60 minutes at RT on a rocking platform at 250 rpm.
It is recommended to incubate cells in a dark moist chamber to avoid fading and drying of the secondary antibody.
- Wash cells three times with PBS for 5 minutes each.
- Add 50 μL of (1:500 diluted in PBS) TOPRO-3 to each slide.
- Wash slides with PBS for 3X 5 minutes.
- Remove excess moisture from slides and add anti-fade solution.
It is recommended not to dry cells completely before adding anti-fade solution.
- Coverslip, seal with nail varnish and leave slides overnight in the dark at room temperature.
It is recommended to use confocal specific coverslips #1.5 (0.16-19 microns thick) and clear nail polish.
While placing the coverslip on slide, care should be taken to avoid the formation of air bubbles.
Image acquisition
- A Zeiss LSM510 Meta Confocal Microscope is used to acquire images using the standard GFP (green, 488 nm), PI (red 543 nm) and far red (blue 633 nm) lasers.
Images acquired using a confocal microscope reveal better spatial resolution.
Since the size of γH2AX foci may be lower than 0.5 μicrons, it is recommended to acquire images with at least a 0.5 μicron step size along the Z-axis.
A 63 x oil immersion objective lens is preferred compared to either higher or lower magnification. For imaging many cells for foci counting it is preferable to use a scan speed of 8 and image size of 1024 x 1024 pixels. To illustrate the spatial relationship between different nuclear proteins it is recommended to use a scan speed of 6 with image size of 2048 x 2048 pixels.
When imaging from multiple channels a line scan acquisition is recommended compared to a frame scan. This avoids bleaching and better resolution is achieved. Sequential scanning is used to prevent bleed through between channels.
Foci counting
Image analysis and foci counting can be performed using various software packages (inlcuding Metamorph, Image-J and Imaris). The procedure for foci counting using Metamorph is described here.
- Open γH2AX image stacks either directly from the file menu or by using the build number stacks option.
- Go to Process menu and chose stack arithmetic.
- Chose Maximum projection and select the planes to be included and click OK.
- Save the resulting image as a γH2AX Tiff. file and open the TOPRO-3 (blue) image.
- Go to regions and select a region drawing tool and draw regions around nuclei.
- Click on γH2AX Tiff image and go to process menu and select morphological filters.
- Chose Top-Hat and select γH2AX image as a source image.
- Apply Top-Hat filter and resulting image will be a binary image with less noise and reduced variation in foci intensity.
Care should be taken when choosing Top-Hat values. Optimal values can be obtained by visual comparison of images before and after applying filters.
- Go to the process menu and select threshold image.
- Choose inclusive threshold and select the lower and higher threshold values.
It is typically the threshold values that affect foci number. Therefore, much attention is required when selecting threshold values. This can be best achieved by the counting foci number using different threshold values in cells exposed to lower than 2 Gy of γ-radiation and then compare foci numbers with manual counting (by eye). The optimal threshold value is the one that minimizes the inclusion of background and maximizes inclusion of foci.
- Select the TOPRO-3 image and go to the regions menu and select the transfer regions option. Select the source image as TOPRPO-3 and the destination as Top-Hat and click OK.
- Once the nuclear regions are transferred to the Top-Hat image go to the measure menu and select Integrated Morphometry Analysis.
- In the pop-up window select source image- Top-Hat and measure by area.
- Select measure all regions and click on measure to obtain the foci number in all of the corresponding nuclei in the Top-Hat image.
- The foci numbers from each region can be exported into a MS-Excel file by using the log- data option.
3D reconstruction of images
To create a 3D image from a stack using Metamorph:
- Go to the stack menu and choose 3D reconstruction.
- Select the angle of rotation (e.g. 160°, 320°).
- Select the 3D reconstruction type- maximum.
- Chose the plane of rotation (horizontal or vertical).
- Chose the Z-calibration distance.
It is preferable to use the user specified Z-distance (optimal is 0.5 μicrons).
- Chose OK to create a 3D reconstruction.
Line scan analysis
For line scan analysis using Metamorph:
- Open either a single plane image or a stacked file.
- Go to the measure tool bars command and select line scan analysis.
- Draw a line across the cell and this will automatically reveal the fluorescence intensity and distance of each marker along the path of line.
To elucidate the spatial relationship of various chromatin markers, for example, γH2AX foci and histone methylation, it is preferable to draw lines which span regions that are both dense and poor in these markers.
Representative Data
For the purpose of this demonstration we used antibodies to γH2AX foci and to H3K4me, to evaluate the spatial distribution of DSBs to an epigenetic determinant of actively transcribing euchromatin. As shown in figure 1, following irradiation with 2Gy, γH2AX foci formed predominantly in regions that were dull for both H3K4me staining and for the DNA stain TOPRO-3 (brightly stained DNA regions are indicative of heterochromatin).
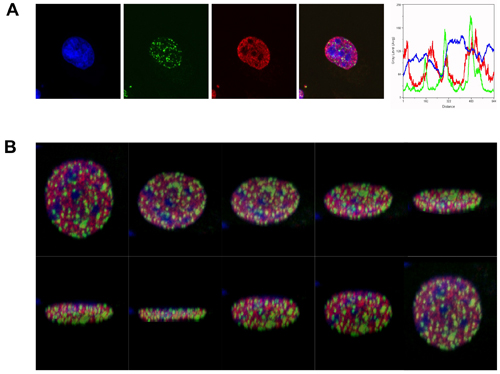
Figure 1. γH2AX foci form predominantly in euchromatin in response to ionizing radiation. (A) Immunofluorescence visualization of γH2AX foci (green) in human erythroleukemic K562 cells, 1 hour after γ-irradiation (2 Gy). γH2AX foci are shown in relation to H3K4me, representing actively transcribing euchromatin (red). DNA is labelled with TOPRO-3 (blue). The merged image (blue, red and green) demonstrates the exclusion γH2AX foci from heterochromatic regions. The line scan analysis (fluorescence intensity vs distance) indicates the relative distribution of the markers in a single plane. (B) Slices at different planes from the 3D reconstruction of the merged image described above.
Disclosures
No conflicts of interest declared.
Acknowledgements
The support of the Australian Institute of Nuclear Science and Engineering is acknowledged. TCK was the recipient of AINSE awards. Epigenomic Medicine Lab is supported by the National Health and Medical Research Council of Australia (566559). This work is funded by the CRC for Biomedical Imaging Development Ltd, established and supported under the Australian Government s Cooperative Research Centres (CRC) program. LM is supported by Melbourne Research (University of Melbourne) and Biomedical Imaging CRC supplementary scholarships. The support of Monash Micro Imaging (Drs Stephen Cody and Iśka Carmichael) was invaluable for this work.
Materials
| Name | Company | Catalog Number | Comments | |
| Roswell Park Memorial Institute -1640 (RPMI-1640) | Growth medium | Invitrogen | 22400071 | RPMI-1640, pH 7.4 medium supplemented with 10% (v/v) fetal bovine, 2mM L-glutamine, 20μg/ml gentamicin, 20mM (HEPES) N-2-Hydroxyethylpiperazine-N’-2-Ethanesulfonic Acid |
| Fetal Bovine Serum (FBS) | Sigma-Aldrich | F2442 | ||
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A7906 | BSA (1%) is used to block any non-specific antibody binding. Primary and secondary antibodies are diluted in BSA. | |
| PBS (without Ca2+ and Mg2+) | Invitrogen | 17-517Q | ||
| Trypan blue | Sigma-Aldrich | T6146 | Used to distinguish between live and dead cells. | |
| Triton X-100 | Reagent | Sigma-Aldrich | T8787 | Triton X-100 (0.1%) used to permeabilise cells. |
| Paraformaldehyde | Reagent | Sigma-Aldrich | 158127 | Paraformaldehyde (4%) used to fix cells. |
| Mouse monoclonal anti-phospho histone-H2AX antibody | Primary Antibody | EMD Millipore | 16193 | Dilution of primary antibody (1:500), in 1% BSA. |
| HistoneH3 (Mono-methyl K4) | Primary Antibody | Abcam | AB8895 | |
| Alexa Fluor 488 goat anti-mouse IgG (H+L) | Secondary Antibody | Invitrogen | 11029 | Dilution of secondary antibody (1:500), in 1% BSA. |
| Alexa Fluor 546 goat anti-rabbit IgG (H+L) | Secondary Antibody | Invitrogen | 11035 | |
| TOPRO3 | DNA Stain | Invitrogen | T3605 | TOPRO3 is a DNA dye with an Abs/Em of 642/661 nm. DAPI could be used if the confocal microscope is equipped with a 405 nm laser. |
| ProLong Gold | Anti-fade solution | Invitrogen | P36930 | This glycerol based mounting medium must be used with an oil based lense that matches its refractive index. |
| Polylysine slides | Menzel-Glaser | |||
| Coverslips (22x50mm) | Coverslips | Menzel-Glaser | CS2250100 | |
| Tissue Culture Flask, Vented Cap | Culture Flask | BD Biosciences | 353112 | |
| Shandon Cytospin 4 | Thermo Fisher Scientific, Inc. | |||
| Cytofunnels | Shandon, Inc. | |||
| Filter Cards | Shandon, Inc. | 353025 | ||
| Coplin Jar, glass | Grale Scientific P/L | 1771-OG | ||
| Staining Trough | Grale Scientific P/L | V1991.99 | ||
| PAP Pen | Zymed Laboratories, Inc. | 008877 | ||
| Gammacell 1000 Elite Irradiator | Gamma Irradiator | Nordion International Inc. | ||
| Zeiss LSM 510 Meta Confocal | Confocal Microscope | Equipped with 3 lasers: 488 nm, 543 nm and 633 nm. | ||
| Metamorph | Software for Imaging analysis | Molecular Devices |
References
- Rogakou, E. P., Boon, C., Redon, C., Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 146 (5), 905-916 (1999).
- Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 273 (10), 5858-5868 (1998).
- Bonner, W. M. . Nat Rev Cancer. 8 (12), 957-967 (2008).
- Dickey, J. S. H2AX: functional roles and potential applications. Chromosoma. 118 (6), 683-692 (2009).
- Kim, J. A., Kruhlak, M., Dotiwala, F., Nussenzweig, A., Haber, J. E. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 178 (2), 209-218 (2007).
- Cowell, I. G. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2 (10), e1057-e1057 (2007).
- Kinner, A., Wu, W., Staudt, C., &, I. l. i. a. k. i. s., G, . Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36 (17), 5678-5694 (2008).
- Vasireddy, R. S., Karagiannis, T. C., El-Osta, A. gamma-radiation-induced gammaH2AX formation occurs preferentially in actively transcribing euchromatic loci. Cell Mol Life Sci. 67 (2), 291-294 (2010).
- Goodarzi, A. A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 31 (2), 167-177 (2008).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved